Molecular phylogenetics has two key objectives: (1) to elucidate the branching pattern of speciation and gene duplication events, and (2) to date the speciation and gene duplication events with a molecular clock. Molecular phylogenetics is instrumental in the discovery of the three domains of life, in providing crucial evidence for the hypothesis of endosymbiosis for the origin of mitochondria and plastids, and in offering a new perspective in a variety of biological research. I illustrate the success of molecular phylogenetics with a few classic and not-so-classic examples.
Although there is only one phylogenetic tree in Charles Darwin’s book (Darwin, 1859), that tree has proliferated over years and spawned a jungle of mathematics and computational algorithms. This chapter does not plunge you right into this jungle. Instead, it will just share with you a few legends and landmarks that may entice you to see more of the jungle in subsequent chapters.
1.1 GENETIC MARKERS ARE IDEAL FOR GENEALOGICAL RELATIONSHIPS
Molecular phylogenetics uses genetic markers as building blocks. The most fundamental genetic marker is the genome responsible for the manifestation of life in any living entity. These genetic markers have been imprinted a history of life, its origin, and its diversification on earth. Molecular phylogenetics aims to reconstruct this history of life from these genetic markers.
I once attended, as a graduate student at Western in the 1980s, a seminar by Dr. Shiva Singh on genetic markers after his sabbatical. Shiva showed photos of a number of places he had visited during his sabbatical, and nobody knew where they were. Finally Shiva flashed a picture of Eiffel Tower and everyone knew that he was in Paris.
“You won’t get lost if you know the landmarks,” Shiva asserted, “and geneticists won’t get lost if they have genetic markers,” and he proceeded to offer a nice presentation on the development and application of genetic markers in solving practical biological and biomedical problems.
Genetic markers go way beyond science. I came across a story of a farmer and his two sons who lived in Germany in early 1970s. The older son spent most of his time working as a farmer like his father. They are both muscular and robust with copper-colored skin. The younger son, in contrast, disliked manual labor. He was not muscular, had pale skin, and did not look quite manly. The morphological difference between the German farmer and his younger son was so obvious that the father decided to go to court to disinherit his younger son, believing that the boy must have resulted from an extramarital affair. At that time, there was no DNA available, but the method of allozyme electrophoresis and immune responses allowed forensic scientists to reach a rather dramatic and surprising conclusion. The younger son was definitely the biological son of the German farmer, but the older one was quite doubtful. The story highlighted how lost the German farmer was without the guidance of genetic markers.
In retrospect, we can see that the morphological similarity between the German farmer and his older son is a consequence of morphological convergence resulting from working hard in the farm. Such similarities are weak for tracing human relationships. Other examples of convergence include the morphological similarities between certain placental mammals and their marsupial counterpart, e.g., between the placental wolf (Canis lupus) and the marsupial Tasmanian wolf (Thylacinus cynocephalus), between the placental cat (Felis catus) and the marsupial tiger quoll (Dasyurus maculatus), and between the placental mouse (Mus musculus) and the marsupial fat-tailed mouse opossum (Thylamys elegans). All these examples of morphological convergence that are not related to true genetic affinity, together with the peculiar phenomenon of mimicry, remind us of nature’s tendency to hide true geological relationships from us.
Genetic markers have been used not only to identify paternity in humans but also in studying multiple paternity in animals. Circumstantial evidence suggests that the white-footed mouse, Peromyscus leucopus, may be promiscuous because (1) males do not provide any sort of parental care (Xia and Millar, 1988) based on observations in semi-natural enclosures, and (2) males gather around females in the field only when females are in estrus (Xia and Millar, 1989). When pregnant females were brought back and allowed to give birth to her young, and when the genotype of both mother and offspring were assessed, one obtains clear evidence of multiple paternity in single litters (Xia and Millar, 1991). For example, a mother’s genotype at one autosome locus is AA, but three offspring genotypes are AA, AB, and AC. This implies that alleles A, B, and C are from males. Because each male has only two alleles, at least two males must have contributed to the litter of offspring.
Genetic markers can also contribute to resolving national conflicts. Between Canada and the United States, there has been a long-term dispute on the management and harvest of Pacific salmon, in particular the allocation of fishing quotas (Emery, 1997). The most fundamental principle of allocation, the equity principle, is to “ensure that each country receives benefits equivalent to the production of salmon originating in its waters.” This principle, rational in its articulation, has one major difficulty in its implementation. That is, how would one know if a salmon caught in the Pacific originated in Canadian or US waters? Fish biologists in the past have studied differences in morphological characters, parasite loads, and many other traits of salmon sampled in Canadian and American rivers, but these traits provide poor resolution for discrimination. Fortunately, sea-type salmons are philopatric and migrate to their natal place to breed. This implies genetic differentiation among salmon populations between Canadian and US river systems. Identification of salmon at species, population, or even individual level is now possible with well-developed DNA markers (Beacham et al., 2017).
1.2 SUCCESS STORIES IN THE APPLICATION OF DNA AND RNA AS GENETIC MARKERS
There are many success stories in the application of molecular phylogenetics, some well-known and some little known. I will present two well-known stories as well as two little known ones, partly because of my conviction that many biological Cinderellas deserve a better fate in real life, and partly because all these stories illustrate the unique insights we can gain only through molecular phylogenetics.
1.2.1 THE DISCOVERY OF THREE KINGDOMS OF LIFE
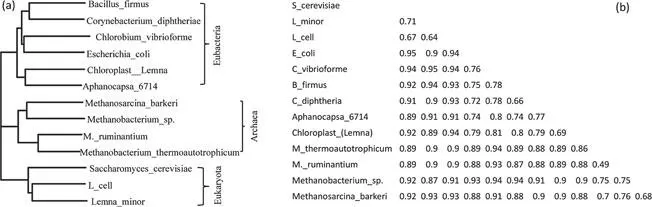
One of the landmark discoveries in molecular phylogenetics is the discovery of three domains of life (Eubacteria, Archaea, and Eukarya) in 1977. Prior to that discovery, we have two domains, prokaryotes without a cell nucleus and eukaryotes with a nucleus. Based on a similarity index (SAB) derived from sharing of RNA fingerprints between taxa A and B, with SAB = 2NAB/(NA+NB), Woese and Fox (1977) showed that the three domains of life are roughly of equal distance from each other. No phylogenetic tree was constructed in that paper, but one can readily derive distances (D) from SAB values and reconstruct a tree (Fig. 1.1a). As the maximum of SAB is 1, we may simply have DAB = 1 – SAB. I have replicated such a distance matrix in Figure 1.1b. The resulting distance matrix, when analyzed by a distance-based phylogenetic method such as FastME (Desper and Gascuel, 2002; 2004) which is also implemented in DAMBE (Xia, 2013b; 2017a), would generate the tree in Figure 1.1a with the representatives of the three kingdoms.
While the result in Figure 1.1 by itself is not strong evidence for the three-kingdom trichotomy, it is sufficient to stimulate further investigation by scientists. This has eventually resulted in empirical substantiation of the three-kingdom classification. In today’s world with almost every corner of the earth being accessible by human, it would have been quite remarkable to discover just a single new species. Imagine how electrified biologists were when a whole new domain of life was discovered!
The effort to trace history back to its origin has gradually shaped a new scientific consensus of cenancestor (Xia and Yang, 2013), the common ancestor of all forms of life. The cenancestor is neither a single cell nor a single genome, but is instead an entangled bank of heterogeneous genomes with relatively free flow of genetic information. Out of this entangled bank of frolicking genomes arose probably many evolutionary lineages with horizontal gene transfer gradually reduced and confined within individual lineages. Only three of these early lineages (Archaea, Eubacteria, and Eukarya) have known representatives survived to this day.
1.2.2 ORIGIN OF MITOCHONDRION AND PLASTIDS (e.g., CHLOROPLASTS)
One of the most fundamental questions in evolutionary biology is the origin of species, but the origin of species ultimately involves the origin of new traits, especially landmark traits such as the origin of mitochondria and chloroplasts. Mitochondria are powerhouses in eukaryotic cells, and chloroplasts allow life on earth to harvest the energy of the sun. How did eukaryotes gain these fantastic organelles?
The endosymbiosis theory, originally proposed by the Russian botanist Konstantin Mereschkowski (1905) but promoted most vigorously by Lynn Margulis (Lynn Sagan) since 1967 (Margulis, 1970; Sagan, 1967) stipulates that mitochondria and plastids in eukaryotic cells represent formerly free-living prokaryotes engulfed by other prokaryotes and reduced in the process of endosymbiosis, around 1.5 billion years ago. However, there was no direct evidence supporting the theory when Margulis articulated the theory, and her paper was rejected about 15 times before its final appearance in the Journal of Theoretical Biology (Margulis, 1995).
The most convincing evidence supporting the endosymbiosis theory came from phylogenetic analysis of conserved segments of small subunit (ssu) ribosomal RNA (rRNA) sequences (Gray, 1989a; 1989b; 1992; 1993) from bacteria, archaea, mitochondria of plants, fungi and animals, and chloroplasts of plants. Mitochondrial sequences from eukaryotic species appear to be monophyletic and their common ancestor clustered with Alphaproteobacteria. Similarly, chloroplast sequences clustered with cyanobacteria. The significant sequence homology represents undisputable coancestry between mitochondria and Alphaproteobacteria, and between chloroplasts and cyanobacteria.
The mitochondrial genome (mtDNA) that perhaps best represents the ancestral state is that of Reclinomonas americana, a heterotrophic flagellate. R. americana has a large mtDNA of 69,034 bp and 97 genes, including 4 genes specifying a multisubunit eubacterial-type RNA polymerase. Almost all extant mtDNA lineages can be viewed as containing a subset of its genes. It is reasonable to infer that the proto-mtDNA is very similar to that of R. americana, and that extant mtDNA lineages were subsequently derived from this proto-mtDNA and now exist as various degenerated forms.
The phylogenetic relationship between the mtDNA in R. americana and bacterial species can be reconstructed by using small and large subunit of rRNA. rRNA genes have been termed universal yardstick (Olsen and Woese, 1993) because they are shared among all living organisms and therefore can facilitate the quantification of their phylogenetic relationship. I have reconstructed such a tree based on aligned ssu rRNA from R. americana mtDNA and from the genome of a diverse array of bacterial species (Fig. 1.2). It is interesting to note that R. americana mtDNA forms a monophyletic group with Rickettsiales, an order of bacteria that are intracellular endosymbionts or pathogens of eukaryotic cells. They all exhibit genome degeneration due to the endosymbiont or parasitic lifestyle. Thus, a mitochondrion is just an extremely degenerated intracellular endosymbiont.
While the original hypothesis of mitochondrial origin by endosymbiosis does not preclude multiple mitochondrial origins through multiple endosymbiotic events, the common consensus is that the protomitochondrion originated only once, through the internalization of a Rickettsia-like bacterium into a host cell which is more likely a prokaryotic cell than a eukaryotic one (Gray, 2012; Lane and Martin, 2010). This ancestral host with the protomitochondrion subsequently diverged into numerous eukaryotic lineages. Only the R. americana lineage still has a mitochondrial genome retaining many of the protomitochondrial states.
I should make a point here that a biologically appealing hypothesis, such as the endosymbiosis hypothesis for the single origin of mitochondria, is often accepted without rigorous and critical examination of relevant evidence. rRNA genes, while universal, may have difficulty even for resolving shallow phylogenies such as vertebrate phylogeny (Xia et al., 2003a). If we replace the mitochondrial ssu rRNA sequence from R. americana in Figure 1.2 by mitochondrial ssu rRNA sequences from other species, we do not consistently observe these other mitochondrial ssu rRNA sequences clustering with species in Rickettsiales. In fact, most of them cluster with bacterial species remotely related to Rickettsiales, that is, evidence for monophyly of mitochondrial rRNA genes is extremely weak. We thus have at least two hypotheses. First, mitochondrial DNA, after accumulating so many substitutions eroding phylogenetic information, is no longer good genetic markers for reconstructing a deep phylogeny. Accepting this hypothesis would effectively rescind the phylogenetic support for the endosymbiosis hypothesis. Second, the diverse array of mitochondria, with many associated genetic codes, re...