![]()
Section 1
Plant Immune Response Pathways
![]()
1
Innate Immunity: Pattern Recognition in Plants
Delphine Chinchilla and Thomas Boller
1.1 Pattern Recognition through MAMPs (Microbe-Associated Molecular Patterns)
Classic work attempted to define and characterize the so-called “elicitors,” pathogen-derived molecules that would elicit a defense response in plants (Darvill and Albersheim, 1984; Boller, 1995). In the case of oomycetes and fungi, these “elicitors” turned out to be characteristic microbial structures derived from their cell walls, such as the heptaglucan epitope of Phytophthora megasperma (see Darvill and Albersheim, 1984) and chitin fragments (Felix et al., 1993), or from microbial membranes, such as arachidonic acid (Preisig and Kuc, 1985) and ergosterol (Granado et al., 1995). Thus, plants appeared to perceive microbes through common patterns that were not specifically associated with pathogens (Boller, 1995). However, although these elicitors were able to induce a vigorous defense response, their importance for actual plant–pathogen interactions remained elusive.
The appreciation of these “general elicitors” changed when a similar principle was described in the field of (human) immunology: In the evolutionarily ancient “innate immunity,” a group of receptors named “pattern recognition receptors (PRRs)” was found to recognize conserved molecular patterns of microbes that are essential for their survival, the so-called “pathogen-associated molecular patterns (PAMPs)” (Medzhitov and Janeway, 2000, Janeway and Medzhitov, 2002). Interestingly, both plants (Gómez-Gómez and Boller, 2000) and animals (Hayashi et al., 2001) were found to possess specific PRRs for bacterial flagellin, namely flagellin sensing 2 (FLS2) and Toll-like receptor 5 (TLR5). This highlighted the similarities of plant and animal innate immunity (Asai et al., 2002), particularly because it appeared that the two PRRs had arisen by convergent evolution rather than from a primeval eukaryotic PRR (see Ausubel, 2005; Boller and Felix, 2009). As well as illustrated by the PRRs for flagellin, it is apparent that the molecular patterns recognized are characteristic of whole classes of microbes, independent of whether they are pathogenic or not, and therefore should more precisely be called “microbe-associated molecular patterns (MAMPs),” a term we will use throughout this chapter (see also Mackey and McFall, 2006; Boller and Felix, 2009; Boller and He, 2009). In fact, well-adapted pathogens might alter and camouflage the molecular patterns that lead to recognition by the PRRs, as illustrated by the changes in the flagellin genes of some plant pathogenic bacteria, such that true pathogens may no longer present the MAMP in question (Felix et al., 1999; Pfund et al., 2004; Sun et al., 2006).
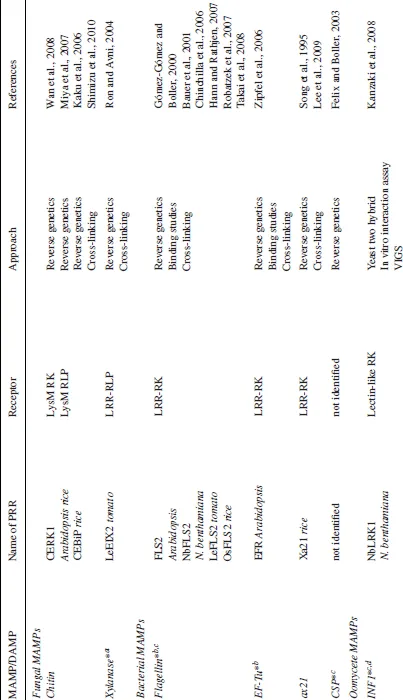
The variety of MAMPs is large, as summarized in Table 1.1. Typically, for a given class of microbes, a given plant species can perceive several different MAMPs. This redundancy guarantees a robust recognition of the microbe.
Table 1.1 Overview of PRRs/binding sites for MAMPs/DAMPs characterized in plants.
While MAMPs are generally characteristic of a whole class of microorganisms, many of these MAMPs are not perceived in a general way by most plants, but only by a few of them, e.g., by most members of an order or family (reviewed in Boller and Felix, 2009). For example, recognition of the active epitope of elongation factor-Tu (EF-Tu), called elf18, is restricted to the Brassicaceae family (Kunze et al., 2004, Zipfel et al., 2006) and bacterial cold shock protein (CSP) is active only in Solanaceae (Felix and Boller, 2003). From an evolutionary point of view, it is probable that perception of EF-Tu and CSP are more recent systems than the perception of flagellin, which is common to numerous plant species (Albert et al., 2010a). There may be an advantage for a given host plant, in terms of coevolution, to recognize a MAMP that most other plants do not.
1.2 Some Classical MAMP-Receptor Pairs
1.2.1 MAMP Receptors: PRRs
In animals, PRRs can be separated into surface-located receptors, called Toll-like receptors (TLRs) and intracellular receptors of the NOD-like family (Takeuchi and Akira, 2010). In plants, the PRRs identified so far are all located at the plasma membrane (Zipfel, 2008). There is currently no example of intracellular recognition of a MAMP in terms of the above definition, although plants possess specialized intracellular receptors of the NOD-like family to perceive effectors (see Chapter 2).
Most plant PRRs described so far belong to the class of receptor-like kinase (RLKs) (Shiu and Bleecker 2001; Shiu et al., 2004): these proteins contain an ectodomain probably acting as the binding site for the respective ligand, followed by a single pass transmembrane domain and a cytoplasmic protein kinase domain, which is likely to function in intracellular signal transduction. Many RLKs are induced by biotic stresses, including MAMP treatments (Zipfel et al., 2004, 2006; Kemmerling et al., 2007), and some were shown to be dispensable for plant development and thus are good candidates as PRRs (Lehti-Shiu et al., 2009).
Three typical leucine-rich repeat (LRR)-RLKs acting as PRRs (which can be referred as LRR-receptor kinases) are FLS2 and EFR, the Arabidopsis receptors for flagellin (Gómez-Gómez and Boller, 2000) and EF-Tu (Zipfel et al., 2006), respectively, and Xa21, an RLK of rice that has long been considered an unusual “resistance gene” product (Song et al., 1995) but was recently shown to perceive ax21 (Lee et al., 2009), a peptide universally conserved in Xanthomonas oryzae that should be considered as a MAMP (reviewed by Ronald and Beutler, 2010; see also Table 1.1 for further examples).
Other functional PRRs are members of a second class of receptor proteins called receptor-like proteins (RLPs). These proteins have a similar structure to RLKs but lack kinase domains; instead, these proteins often exhibit a short cytoplasmic domain with no signaling signature. This suggests different molecular mechanisms of receptor activation than those controlling RKs; they probably have to interact with other membrane proteins to transmit the signal across the membrane (Wang et al., 2008a). RLPs have a similar structure to the animal TLRs, but in contrast to TLRs that are encoded by 10–12 different genes in mammals (Leulier and Lemaitre, 2008), RLPs expanded into a larger family, with 57 members in the Arabidopsis genome (Wang et al., 2008a). The apparent expansion of the families of RLKs and RLPs may indicate that in evolution these receptors have become one of the preferred systems for non self-perception in plants. To date, two RLPs have been identified as PRRs for individual MAMPs: (1) the receptor for the fungal MAMP xylanase (ethylene (ET)-inducing xylanase, EIX) in tomato, named LeEIX (Ron and Avni, 2004); (2) and the chitin-binding site CEBiP (chitin elicitor binding protein) in rice (Kaku et al., 2006).
PRRs are often identified on the basis of genetics, and only for a small number, biochemical evidence has been provided to demonstrate direct interaction between the receptor and its respective ligand (Table 1.1). However, technical advances in methods, such as affinity chromatography, chemical cross-linking, and immunoprecipitation, have allowed the unequivocal identification of PRR–MAMP interactions and will hopefully shape the future of PRR characterizations (Chinchilla et al., 2006; Kaku et al., 2006; Shinya et al., 2010).
1.2.2 Flagellin Perception in Arabidopsis through FLS2: A Paradigm for MAMP Recognition in Plants
Flagellin, the best-characterized MAMP in plants, was serendipitously identified as the active elicitor in a “harpin” preparation from the plant pathogen Pseudomonas syringae (Felix et al., 1999). Flagellin is the main building block of the flagellum, which allows bacteria to “swim”; typically, a flagellum consists of 10,000 monomers of flagellin. Due to its essential role in bacterial motility, and also due to its abundance and surface exposure, flagellin represents a perfect “molecular pattern” to detect the approach of potentially pathogenic bacteria. The “epitope” of flagellin perceived by plants is located at the N-terminus of the protein and is strongly conserved in bacteria (Felix et al., 1999). Its effect can be mimicked by a synthetic peptide called flg22. Most higher plants, including both gymnosperms and angiosperms, share the capacity to perceive flg22 as a MAMP (Albert et al., 2010a).
Among responses typical for MAMPs (see also next Section 1.3), a prolonged treatment with flg22 induces a growth arrest in seedlings. Using this growth defect as a readout, a genetic screen was conducted in Arabidopsis that allowed the identification of FLS2 as a gene essential for flagellin responses (Gómez-Gómez and Boller, 2000). Using a biochemical approach with radiolabeled flg22, it was shown that these mutants were not only impaired in flg22 signaling but also in flg22 binding (Gómez-Gómez et al., 2001). Both Arabidopsis and tomato perceive flg22, but interestingly these two species show different specificities of binding and responses to derivatives of flg22 (Meindl et al., 2000; Bauer et al., 2001). Transfer of Arabidopsis FLS2 into tomato cells (having an endogenous flagellin receptor) induced new recognition specificities representative of the flagellin receptor from Arabidopsis indicating that AtFLS2 is the “bona fide” receptor for bacterial flagellin, controlling binding of ligand and activation of responses (Chinchilla et al., 2006). This was corroborated by cross-linking experiments with labeled flg22, followed by immunoprecipitation with antibodies specific for FLS2, which showed unequivocally that FLS2 interacts directly with its flg22 ligand (Chinchilla et al., 2006). Several orthologs of FLS2 were identified in tomato (LeFLS2; Robatzek et al., 2007), Nicotiana benthamiana (NbFLS2; Hann and Rathjen, 2007), and rice (OsFLS2; Takai et al., 2008). In rice, responses to flg22 are weak, but OsFLS2 is able to functionally complement Arabidopsis fls2 mutants, confirming that it is indeed a flagellin receptor (Takai et al., 2008). Comparison of orthologs of FLS2 indicated a highly conserved structure for this protein, including a large ectodomain of 28 LRRs, except for OsFLS2 lacking LRR3, and other conserved domains at the N- and C-terminus, indicating in particular that the large LRR domain is of functional relevance (Boller and Felix 2009).
It remains an open question where the exa...