![]()
1
Defining the equine genome: The nuclear genome and the mitochondrial genome
Bhanu P. Chowdhary
A genome represents the genetic material or the core regulatory and functional machinery of any organism. In the horse, as is typically the case in most mammals, the genetic material largely resides in the nucleus of the cell, while a fractional yet functionally vital component is present in the cytoplasm. The former is packaged in the form of chromosomes and the latter in the form of mitochondria. It is difficult to assign greater significance to one over the other, hence a debate in that realm has to be left at accepting that the genes contained in each of the components have an important role to play, individually as well as in conjunction with other genes in a network or pathway, bringing wholeness to the functioning of the horse.
This chapter first provides an overview of the nuclear genome in terms of chromosome number, standard karyotype, and chromosome nomenclature, and highlights basic facts regarding equine chromosomes. The description includes a brief introduction to structural and chromosomal aberrations and their impact on overall viability of horses. Finally, a summarized overview of the current knowledge of the equine mitochondrial genome is presented. The information serves as a foundation and a reference point to all aspects of the equine genome presented and discussed in relation to mapping, genetic variation, phenotypic variations, diseases, and other such information in subsequent chapters.
Nuclear Genome of the Horse
Chromosome number, karyotype, and schematic presentation
The size of the nuclear part of the equine (Equus caballus; ECA) genome is around 2.7 megabase pairs (Mbps). The nuclear genome is packaged in 64 chromosomes (diploid chromosome number presented as 2n = 64) that may be metacentric, sub-metacentric, or acrocentric. It is noteworthy that the genus Equus has 8 extant species and the chromosome number in these species varies from 2n = 32 in the Hartmann's mountain zebra to 102 in the Domestic ass and the Mongolian and Transcaspian wild asses, although marked similarities in size and gene content exists between them (Chowdhary & Raudsepp, 2000). The 64 horse chromosomes are comprised of 32 pairs of homologues (one coming from sire/stallion and the other from the dam/mare), of which 31 pairs are autosomes that have an identical gene set present essentially in the same order from one end to another, and one pair is the sex chromosomes that are similar in females (two X chromosomes) but different in the males (one X chromosome and one Y chromosome). The X-chromosome is the second-largest element among the equine chromosomes and forms about 5% of the genome, whereas the Y is one of the smallest (Chowdhary & Raudsepp, 2000).
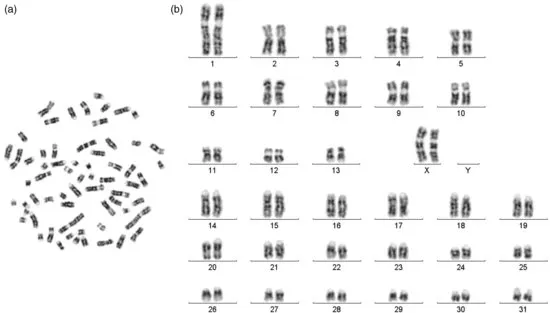
Horse chromosomes have now been studied under the microscope for more than a century. Though Makino (1942) established that, like other mammals, horse also has an XY sex chromosome system, the correct chromosome number in horses – and the confirmation of this number by independent groups – became known much later (Rothfels et al., 1959; Moorhead et al., 1960; Makino et al., 1963). The 14 pairs of metacentric and sub-metacentric horse chromosomes, including the sex chromosomes and the 18 pairs of acrocentric autosomes, are arranged in a specific pattern (referred to as the karyotype; see Figure 1.1) for a quick visual assessment of the chromosomes. The arrangement has evolved over the past several decades through iterations of internationally agreed “standardizations” of which the last, and potentially the final, one was carried out by the International System for Chromosome Nomenclature of the domestic Horse (ISCNH, 1997). This standardized karyotype gave a detailed description of both G- and R-banded chromosomes along with schematic drawings (idiograms) of individual chromosome pairs showing landmarks and band numbers for each of the key banding approaches. Three size resolutions ranging from moderate to reasonably elongated chromosomes are provided to facilitate easy identification of the chromosomes. Overall, the current standard karyotype serves as a basis of “cross-talk” on individual horse chromosomes among equine cytogeneticists and researchers worldwide.
Application of different banding techniques
A variety of staining/banding techniques have been applied to analyze equine chromosomes. The ultimate goal of using these banding techniques was to exploit a range of structural and functional features that permit unambiguous discrimination of individual pairs of homologous chromosomes. The dyes used to stain the chromosomes can be fluorescent or nonfluorescent. The most common staining technique for visualization of chromosomes is the use of Giemsa (Trujillo et al., 1962). The Q-banding (Caspersson et al., 1970) and G-banding (Seabright, 1971) techniques highlight AT-rich DNA as positive bands. The R-banding technique (positive bands are reverse to the G- and Q-positive bands) primarily highlights GC-rich DNA by fluorescent (RBG; Molteni et al., 1982) as well as heat treatment/Giemsa-staining-based approaches (RHG; e.g., Power, 1987). These bands can be enhanced by obtaining elongated chromosomes via using cell cycle synchronization and incorporation of bromodeoxyuridine (BrdU) into chromosomal DNA (e.g., Romagnano et al., 1983; Marki and Osterhoff, 1983; Power, 1987a, 1990) as done by Rønne et al. (1993). Essentially, bands are like bar codes that are unique for a chromosome pair among the pairs of homologous chromosomes in a species, which allows distinguishing the homologues from the set of chromosomes in a metaphase spread.
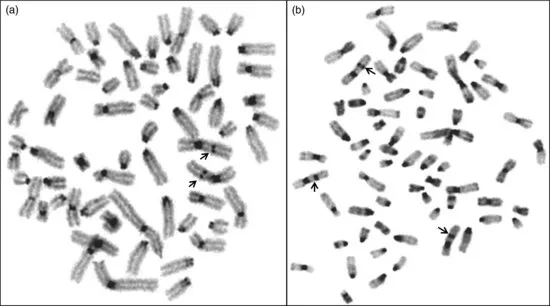
Some banding techniques like the C-banding specifically depict constitutive heterochromatin. The “bands” or regions are highlighted by acidic and/or alkaline and heat treatment, and are seen on almost all horse chromosomes, except ECA11. Interstitial heterochromatin is reported on ECA1pter, 12q, and Xq (most prominent), whereas the Y-chromosome is known to be largely heterochromatic (Figure 1.2). T-banding, which highlights telomeric repeat sequences present at their usual terminal location of the chromosome with no intercalary presence, is observed in horses. They were first reported using molecular cytogenetic approaches (de La Seña et al., 1995). Next, regions containing transcriptionally active ribosomal RNA genes (rDNA or nucleolus organizing regions, NORs) are visualized by NOR-banding carried out by staining chromosome preparations with silver nitrate (Goodpasture and Bloom, 1975). Molecular approaches allow identification of both active and inactive regions in a cell. Typically, ECA1, 27, 28, and 31 are considered as the NOR-bearing chromosomes. In addition to these banding techniques, attempts have also been made to produce bands on equine chromosomes specific for electron microscopy (Messier et al., 1989; Richer et al., 1989), which has even allowed the construction of a karyotype.
Chromosome aberrations – a brief overview
A vast array of reports on chromosomal abnormalities shows that equine chromosomes have been fairly extensively studied, even though the number of published reports is substantially lower than in cattle and pigs. As in other mammals, these chromosomal aberrations are classified into two major categories, namely (1) autosomal (includes autosome-sex chromosome and vice versa translocations) and (2) those involving only the sex chromosomes. Each of the categories is further classified into two subheads: structural and numerical. A large number of mosaic/chimeric karyotypes involving the sex chromosomes, as well as cases of sex-reversal, have been reported. We estimate that not more than 3,000 horses (predominantly mares) have been examined cytogenetically, resulting in the detection of about 400 cases with chromosome aberrations. This count does not include about 120 cases of sex reversal. As the majority of these cases were hand-picked, either due to phenotypic deviations or due to fertility problems, no proportionate evaluation of the frequency of chromosomal aberrations in the horse could be projected.
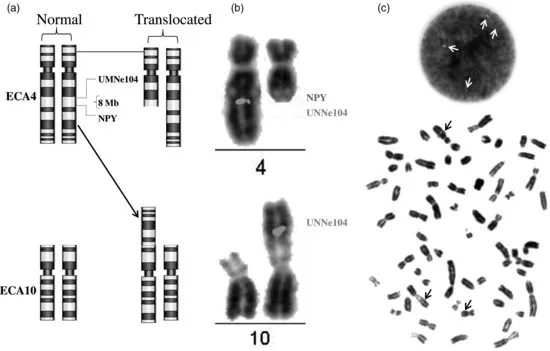
Rather few structural aberrations have been detected in horses. These include deletions (loss of part of a chromosome), translocations (both balanced and unbalanced), inversion, and duplication. Figure 1.3 shows one of the most recently discovered balanced translocations in the horse that has been analyzed using traditional and molecular techniques (Das et al., forthcoming). All numerical aberrations in horse are trisomies of different autosomes, all of which involve small acrocentric chromosomes (nos. 23–31). Some of the salient features of viable individuals with autosomal aberrations (both structural and numerical) are: reduced to completely impaired fertility, minor to moderate anatomical malformations primarily affecting gait or orientation, atypical poor confirmation, and moderate to notably smaller height as compared to the breed/parents average. Like in other species, production of offspring to full term by individuals bearing balanced reciprocal translocations and tandem fusions has been reported in the horse (see Power, 1991; Long, 1996; Durkin et al., forthcoming; Das et al., forthcoming); still, invariably varying degrees of reduced fertility is observed. However, foaling of an ECA26 trisomy carrier to produce a karyotypically normal colt (Bowling and Millon, 1990) is noteworthy.
The vast majority of chromosomal abnormalities reported in the horse involve the sex chromosome (predominantly the X). Of the approximately 350 abnormal karyotypes reported to date, more than 90% involve the sex chromosomes. The types of structural chromosome abnormalities involving the sex chromosomes vary from translocations and deletions to isochromosomes, invariably all impacting fertility. A wide range of numerical aberrations of the sex chromosomes are known in the horse, of which monosomy (lack) of the X chromosome (karyotype constitution 63,XO) is the most common. Few cases of pure trisomy of the X chromosome (65,XXX) have also been reported in the horse. Typically, XO and XXX mares are infertile. Next, cases of pure 65,XXY (Kubien et al., 1993) and 66,XXXY (Gluhovschi et al., 1970, 1975) have also been reported.
Finally, various categories of mosaics/chimeras involving the sex chromosomes (primarily the X) have hitherto been reported in horses. It is noteworthy that most of the reported chimeric/mosaic cases are registered as females with a wide range of clinically detectable reproductive system deviations. The animals show varying degree of virilization of the external genitalia.
Sex reversal, most appropriately described as a disagreement between the karyotypic sex and the phenotypic/anatomical sex, has been described in horses. Approximately 135 such cases have to date been observed. Normally, individuals with XY sex chromosome constitution are expected to be males (64,XY). Sex reversal is a condition in which the aforementioned individuals appear more like females, with varying degrees of male-like characteristics. The terminology holds even if the karyotype indicates the animal to be a female (64,XX), but phenotype and clinical examination show the preponderance of male-like characteristics. Some of these cases and associated molecular work we carried out to analyze these cases in relation to the observed micro-deletions on the horse Y chromosome are described in Chapter 5.
Mitochondrial Genome of the Horse
Structure, function, and utility
The mitochondria (mt) are the powerhouses of cells and are responsible for more than 90% of mammalian energy production. All aerobic respiration within cells occurs in this organe...