![]()
1
The Background
1.1 Meet the Cast
The main point about microbes is that they are very small. Their one unifying feature is that they are too small to be seen without the aid of a microscope – although even that definition, as we will see later on, is blurred at the edges, as some of the ‘microbes’ I will consider are actually quite big. Within the basic definition, there is a substantial range of diversity. You will probably have heard of some of these microbes, such as the influenza virus or the bacterium known as MRSA, as they regularly make the news because they inflict themselves on us. Others may also be familiar because of their everyday role in fermentation – think of the yeasts that are needed for the production of bread, wine and beer, and the ‘friendly’ bacteria that are used for yogurt making. As we will see later on, there are very many more examples of a wide range of microbes that are of direct importance to us, both in disease (and health) and economically.
But this is only the tip of an enormous iceberg. Microbes are all around us, in vast numbers and diversity, especially in soil and water. It has been estimated that the world has 1031 bacteria – that's 1 with 31 zeros after it – with a total biomass greater than all the plants and animals combined (and that is just the bacteria, before we add in the other microbes – viruses, fungi, algae and protozoa). They play a massive role in shaping our environment, including fixing carbon and nitrogen from the air – and, by degrading organic matter, in releasing these elements again into the air. Our knowledge of the diversity of microbes in the environment has increased enormously in recent years. Molecular techniques that we will encounter in a later chapter have shown that most (perhaps 99 per cent) of these organisms were previously totally unknown and have never been grown in the laboratory.
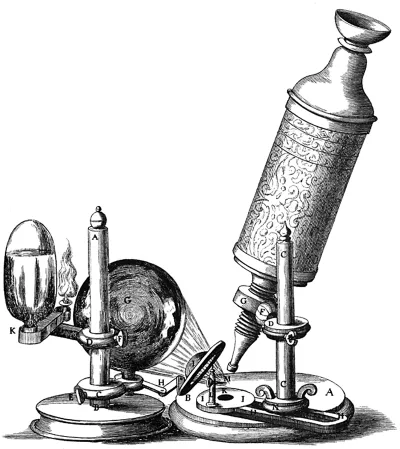
We can begin the story in the 17th century, in Holland. Antonie van Leeuwenhoek was born in Delft, in 1632. After an apprenticeship with a cloth merchant, he set up his own drapery business and became a prosperous and influential citizen of Delft. Having seen the magnifying glasses used by textile merchants for examining the cloth, he developed an interest in the use of lenses and started to make his own as a hobby. Although his ‘microscopes’ were simple by modern standards – consisting of just a single lens – the superb quality of his lenses, and his skill and patience in using them, enabled him to make many important observations. These included the first descriptions of microscopic single-celled organisms (which he called ‘animalcules’), which he reported to the Royal Society in London in 1676.
These observations met with a considerable amount of scepticism but, eventually, after much further investigation, his achievements were recognized and he became a Fellow of the Royal Society in 1680. He continued to make many further detailed observations, such as the description of bacteria in plaque from teeth, until shortly before his death in 1723. Unfortunately, he kept secret some of the crucial details as to how he made his lenses, so, with his death, that part of the story came to an end.
However, others had also developed and used microscopes at around that time. Robert Hooke (1635–1703) is perhaps best known today for his study of the elasticity of materials, described by the mathematical relationship we still know as Hooke's Law. But that was far from the total of his interests or achievements, which ranged from experimental science to architecture. The part of his work we are concerned with here was his role in the development of the compound microscope (which, like a modern microscope, contained two lenses rather than the single lens used by van Leeuwenhoek).
He used this microscope to make a large series of observations of diverse biological materials, which was published in 1665 as a book, Micrographia. Notably, his description of the microscopic structure of slices of cork was the first identification of the cellular structure of, in this case, plant material (he coined the word ‘cell’ for them because of their resemblance to cells in a monastery). Although the microscopes used by Hooke (and other similar ones of that period) were more like a microscope of today than van Leeuwenhoek's single lens instruments, the technical difficulty of making them, and the superb craftsmanship of van Leeuwenhoek, meant that they were actually inferior to van Leeuwenhoek's.
It's now time we met the cast so, for the first members, let's consider viruses. These are so different from other microbes that it is only a matter of convenience that we do include them as ‘microbes’. Indeed, it is questionable as to whether we should consider them as ‘living’ at all (I'll come back to that question in Chapter 10).
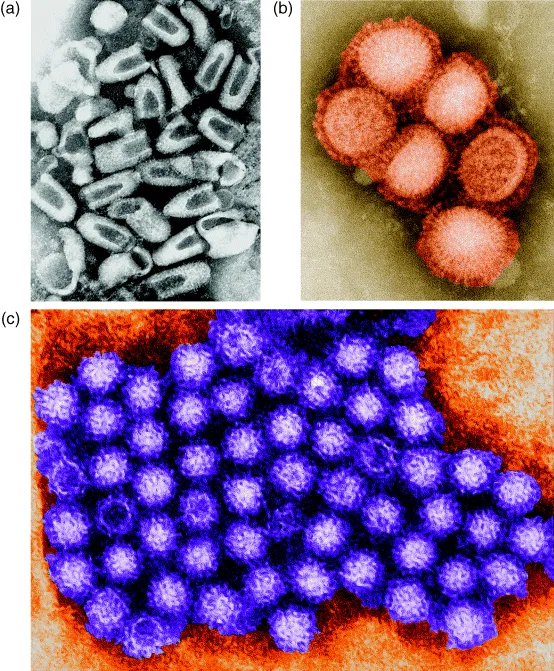
Viruses are not able to replicate, or to do anything at all, outside a host cell. The simplest viruses consist just of a nucleic acid molecule (which can be RNA or DNA, but not both), surrounded by a protein coat. These contain a limited number of genes. For example, one of the most basic viruses, called MS2, which infects E. coli, has just three genes: one codes for the coat protein, one is needed for copying the viral genome (RNA in this case) and the third is used to organize the assembly of the virus particle.
However virus structures, and sizes, are quite diverse. Many, including some important human pathogens, are much larger than MS2 and have complex structures including, in some cases, a lipid coat. But virtually all viruses are so small that the human eye cannot see them, even with the aid of a light microscope –an electron microscope is needed to ‘see’ them. There are, however, some that are larger. The largest known viruses, such as the mimivirus which infects protozoa, are similar in size to some of the smallest bacteria, and they have a genome size to match. But even the largest and most complex viruses are completely unable to replicate without infecting a suitable host cell.
At several points within this book we will consider viruses that infect bacterial cells. These are called bacteriophages, or just phages for short (the word ‘phage’ being derived from the Greek ‘phagein’, meaning ‘to eat’). These are very widespread in nature and they can be of real practical significance – a phage infecting a bacterium that is used, for example, in the production of yoghurt can cause serious economic losses. But they have also played a major role in scientific research. Much of our knowledge of how genes work comes originally from studies of phages, where the simplicity of their structure, and the ease with which they can be grown and manipulated, made them invaluable models.
When a phage infects a bacterial cell, its nucleic acid (DNA or RNA, depending on the phage) is injected into the cell. Some of its genes are then recognized by the cell's machinery, which obligingly makes the relevant proteins that those genes code for. These proteins then divert the cell's activity away from its own genes and towards the production of many copies of the phage nucleic acid. At some point in this process, the DNA of the host cell is usually broken down and the bits are used for making the nucleic acid of the virus. The proteins that make up the external structure of the phage (the coat) are then produced, and the nucleic acid is packed into that structure. The consequence of this is the lysis of the bacterial cell and the liberation of hundreds or thousands of copies of the virus. The whole process, from infection to lysis may take perhaps 20–50 minutes (depending on the phage). The details of this process vary considerably from one phage to another, but the general principles are similar. An equivalent, but more complex, process occurs when viruses infect higher (eukaryotic) cells, including human cells.
Although we usually think of viruses as causing diseases, this does not always happen. Some viruses have the ability to remain latent within an infected cell. We may only realize that they are there when the latency breaks down, perhaps due to a drop in our immune defences. This happens, for example, with the herpes virus that causes cold sores, typically around the lips, and the varicella-zoster virus, which initially causes chickenpox but can subsequently remain dormant until causing an outbreak of shingles many years later.
The best understood example of latency is a virus (bacteriophage), known as lambda, that infects E. coli. This has a very elegant mechanism for ensuring that, in a proportion of newly infected cells, the expression of all the virus genes is turned off, apart from one gene that codes for a protein that is responsible for maintaining this repression. In this state, known as lysogeny, the DNA of the virus is integrated into the chromosome of the host cell and is therefore copied, along with the rest of the DNA, as the bacterial chromosome is copied during growth. Each daughter cell therefore receives a copy of the virus DNA. Studies of genome sequences have revealed that most bacteria (and, indeed, animal cells, including our own) contain a number of copies of a variety of viruses, stably integrated into the chromosome and never showing any signs of their presence.
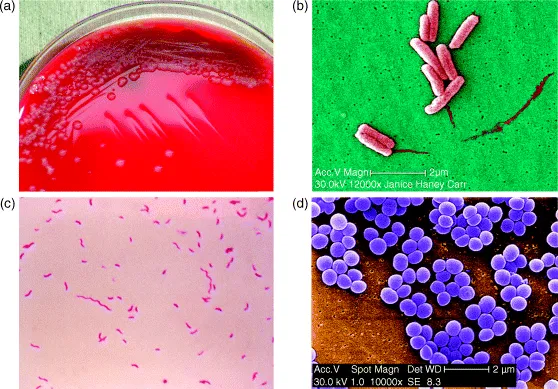
Our second class of microbes is the bacteria and, by way of introduction, I will look at one bacterium in particular: Escherichia coli, or E. coli for short (unfortunately, most bacteria – and many other microbes – do not have simple common names, so we have to get used to using Latin ones). This is described as a rod-shaped organism, but it is better to visualize it as a short cylinder with rounded ends. Later on we will encounter bacteria with other shapes, especially ones such as Staphylococcus, which are spherical, as well as bacteria which grow as filaments or in spiral shapes.
E. coli, which is a common inhabitant of the human gut (as well as being able to cause some nasty diseases), is 2–3 μm long (a μm, or micrometre, is a millionth of a metre, or a thousandth of a millimetre) and 0.5–1 μm wide. It is a favourite model organism for bacteriologists because it will grow readily in a simple medium – all it needs is a sugar such as glucose and a nitrogen source such as an ammonium salt. It will grow even better if it is given a richer medium containing, for example, yeast extract. In a rich medium, it will divide every 20 minutes or so (bacteria typically grow in an apparently simple way – a cell gets bigger until it reaches a certain size, then it divides into two cells, which in turn grow and then divide again – so they multiply by dividing!).
If we start with one cell, after 20 minutes there will be two, after 40 minutes four cells and, by one hour, eight cells. This is known as exponential, or logarithmic growth – it starts off slowly, but very soon reaches astronomical numbers. After ten divisions, there will be about one thousand cells; after another ten divisions, the number will reach a million; after a further ten divisions, it will be up to a thousand million cells.
When we get to such large numbers, they become very difficult to handle in the usual way, so we use what is known as scientific notation (see Appendix 2 for further explanation). A thousand, for example, is (10 × 10 × 10) so we call it 103 rather than 1,000. A million (1,000,000) is 106, and a thousand million (1,000,000,000) is 109. So, after 30 cell divisions (about ten hours), we would have some 109 bacteria in our flask. This process does not continue indefinitely of course. After a while, the bacteria start to run out of nutrients (diffusion of oxygen into the medium is usually the first limiting factor for E. coli) and they will stop growing. For E. coli, this will usually happen when the concentration of bacteria reaches about 109 cells per millilitre. In other words, a 5 ml teaspoon would contain five billion, or five thousand million, bacteria.
One practical consequence of these massive numbers is worth a slight digression here. Disinfectant manufacturers will commonly make claims such as ‘kills 99 per cent of household germs’. This sounds impressive – until we consider the numbers involved. If we start with say 106 bacteria (which is not really very high), then killing 90 per cent (or leaving ten per cent remaining) will reduce the numbers to 105; even killing 90 per cent of those (which leaves one per cent of the original), there will still be 104 bacteria. So killing 99 per cent (or leaving one per cent untouched) merely reduces the numbers from 106 to 104 (which we refer to as a 2-log reduction). Even if we kill 99.9 per cent, we still have 103 bacteria. It is a useful effect, but not as dramatic as the original claim sounds.
When we grow a bacterium in a liquid medium (a liquid culture), it goes through several recognizable stages. When the culture is inoculated (that is, a relatively small number of bacteria are put into the broth), nothing much appears to happen for a while. This is the so-called lag phase. Essentially, the bacteria are getting used to the change from the resting state in which they have been stored, and they are responding to the availability of food by making all the various components needed for growth. Some genes (those needed for the resting stage) are switched off, while the genes needed for active growth are switched on. We'll look further at what is involved in these switches in Chapters 7 and 8.
When the cell is ready, it will start exponential growth. At the end of the log phase, when it runs out of food, the process is, in effect, reversed. The genes needed for active growth are switched off, and the cell enters stationary phase. This is not merely the absence of growth. A number of functions are necessary if the cell is to stay alive in stationary phase, so these genes have to be switched on.
Many bacteria, such as E. coli, survive quite well in stationary phase, but not all do. Some will start to die, presumably because they do not have the genes needed to keep the cell alive in the absence of growth. Later on, we will also encounter bacteria that are able to form specialized cells known as spores, some of which can survive almost indefinitely without any detectable metabolic activity. These dormant structures are extremely important in a practical sense, as they may be extremely resistant to heat and disinfection. Examples of spore-forming bacteria include the organisms responsible for tetanus and botulism (see Chapter 5).
The conventional way of identifying bacteria in a mixture – such as might be obtained from a clinical specimen such as a wound swab, or from an e...