![]()
Chapter 1
Introduction
Frans J. de Bruijn
In this second volume of Handbook of Molecular Microbial Ecology, examples are given of metagenomic studies in a large variety of habitats and using diverse techniques. Part 1 discusses the Metagenomics of Viral Genomes, with an Introduction in Chapter 2 and sample Chapters on viruses in soil and aquatic environments, modern strombolites and thrombolites, Yellowstone hot springs, human specimens and plants. In the last case, Chapter 7 proposes next-generation sequencing and metagenomic analysis as a novel, universal diagnostic tool in plant virology. Various methods are described to generate viral metagenomes and to assemble and analyze them.
In Part 2, the soil habitat is the subject, with an Introduction in Chapter 9 and topics addressed include methods for soil DNA and RNA isolation and purification for multiple metagenomics applications (Chapters 10 and 11). These techniques are essential for construction of (large insert) clone libraries (see also Volume I, Chapter 22) and random DNA sequencing studies. New approaches to retrieve full-length functional genes and phylogenetic analysis of bacterial populations or major soil-borne lineages, as well as rare members of the soil biosphere (Chapter 15), using the 16S rRNA gene and metagenomic libraries, are presented. The soil antibiotic resistome is also analyzed (Chapter 17).
In Part 3, chapters on the digestive tract are presented with an Introduction in Chapter 18, which includes the human gut microbiota, and the possible correlation of human disease with human microbiota (Chapters 18–21). Some of these studies are part of consortia such as The Human Microbiome Project, and The Human Gutna Microbiome Initiative, which are discussed in Volume I, Chapter 35. This part is complemented with chapters addressing the metagenomics of termite guts and buffalo rumens (Chapters 22 and 23).
In Part 4, the metagenomics of microbiota in the marine and lake habitats are the subject of study. Microbial diversity in the deep sea, deep sediments and the underexplored “rare biosphere,” as well as lakes is investigated (Chapters 24–31). Genomic adaptations in marine organisms (Chapter 26), the ecological genomics of marine Picocyanobacteria (Chapter 30) and the diversity and role of bacterial integron/gene cassette metagenomes in extreme environments (Chapter 31; see also Volume I, Chapter 26) are highlighted. These studies are complemented by a metatranscriptome analysis of complex marine microbial communities (Chapter 27; see also Volume I, Chapters 62–64).
In Part 5, metagenomic analysis of microbes in a varied number of habitats is presented, ranging from gutless marine worms, human saliva, an acid mine draining environment, terrestrial hotsprings, deep-sea hydrothermal vents, biogas plants, visicomyid host clams, to the surface of building stones (Chapters 32–42). The purpose of this section is to expose the reader to many different habitats and metagenomic approaches to study them.
In Part 6, studies on the application of metagenomics to the discovery of biodegradation genes/enzymes from different habitats, such as aromatic degradation pathway genes, benzoate degradation genes, an alcohol/aldehyde dehydrogenase gene, and alkane hydroxylase genes are presented (Chapters 43–46).
In Part 7, the metagenomic discovery of several novel natural products by different methods is presented. An overview of “functional Metagenomics and its industrial relevance” is presented in Chapter 47. The examples shown include a cold active lipase, coenzyme B12 dependent glycerol dehydratase- and diol dehydrogenases, biomedicals, and antibiotics, as well as the discovery, development and commercialization of Pyrophage 3173 DNA Polymerase (Chapters 48–53).
These parts do not mean to be all inclusive, but should serve the reader with examples of different approaches to use in their own systems/habitats, as well as provide references back to the original publication(s) the chapter was derived from and an extensive literature on the topic.
Volume II ends with a summary section comprising a perspective on the future of the “omics” and single-cell analysis (Chapter 54), as well as an article on the birthday of Darwin's “On the Origin of Species” and the relevance of Darwin's work to today's molecular methods and species concepts (Chapter 55).
![]()
Part 1
VIRAL GENOMES
![]()
Chapter 2
Viral Metagenomics
Shannon J. Williamson
2.1 Introduction
The term “metagenomics” was coined by the soil microbial ecologist Dr. Jo Handlesman in 1998 [Handelsman et al., 1998]. Metagenomics, or community genomics, refers to the study of the genomic contents of microbes extracted directly from the environment. The establishment of metagenomic techniques was an important breakthrough in microbial ecology because microbes that can be cultivated in the laboratory are thought to account for less than 1% that exist in many environments [Whitman et al., 1998]. Over the past decade, metagenomics has developed into an emerging field of study for researchers specializing in diverse disciplines, and metagenomes have been created from the simplest biological complexes (i.e., viruses—the subject of this review) as well as from assemblages of eukaryotes. Metagenomic sequence data are typically used to address the following two fundamental questions: Who is there? and What are they doing? The taxonomic and functional data from metagenomic studies have revolutionized our understanding of the diversity of microbes and the roles they play in their communities.
Viruses are abundant and ubiquitous biological components of every biome on Earth and outnumber all cellular forms of life. Viruses have been the subject of scrutiny for nearly 120 years, but the field of viral metagenomics is relatively young, with the first marine viral metagenome published in 2002 [Breitbart et al., 2002]. Since this time, the number of published viral metagenomes (viromes) has exploded, and the adoption of next-generation sequencing technologies by researchers has resulted in a relative deluge of information on the genomic contents of viral communities inhabiting a diverse range of environments (see Chapter 3 in this volume for a comprehensive list of viromes). Analysis of viromes has enabled a deeper understanding of virus community dynamics including genotypic and taxonomic diversity, functional capacity, biogeography, and evolution. Viral metagenomics is also a powerful tool for viral discovery and has been used in this capacity to reveal the presence of novel DNA and RNA-containing viruses in a variety of samples, ranging from seawater to domesticated plants. This chapter reviews the technical and applied aspects of viral metagenomics, highlighting examples from both “natural” and human-derived environments.
2.2 Experimental Approaches and Sequencing Technologies
Due to their ubiquitous nature, it's possible to collect viruses from almost all types of biological samples. Indeed, viral metagenomic approaches have been applied to samples collected from a diverse range of environments: from marine waters and sediments to the human gut to a vineyard [Breitbart et al., 2002, 2003, 2004a, 2008; Angly et al., 2006; Fierer et al., 2007; Desnues et al., 2008; see also Chapter 5, Vol. II; Kim et al., 2008; McDaniel et al., 2008; Schoenfeld et al., 2008; see also Chapter 6, Vol. II; Vega Thurber et al., 2008; Williamson et al., 2008; Djikeng et al., 2009; Nakamura et al., 2009; Coetzee et al., 2010]. However, the techniques used to extract virus particles vary and are dependent on the type of sample under study. While the isolation and concentration of viruses from aquatic ecosystems is rather straightforward, more complex matrices (such as soils, sediments, tissues, and clinical samples) present a greater challenge. Collection and purification of virus particles is followed by the targeted extraction of viral nucleic acids, either DNA or RNA, with nuclease-mediated destruction of nontargeted molecules. Alternatively, all types of viral nucleic acids (double-stranded or single-stranded DNA or total RNA) can be purified from a sample simultaneously using hydroxyapatite chromatography [Andrews-Pfannkoch et al., 2010].
Amplification of viral DNA is often performed in order to (1) provide sufficient quantities of nucleic acid for library construction and sequencing, (2) produce unmodified copies of viral DNA [Breitbart et al., 2002], and (3) purify the DNA of potential contaminants that may interfere with downstream molecular applications [Thurber et al., 2009]. Amplification can be accomplished using a linker-mediated approach [Andrews-Pfannkoch et al., 2010] or by multiple displacement amplification using the phi29 DNA polymerase [Thurber et al., 2009]. Lastly, depending on the sequencing technology to be employed, clone-dependent or clone-independent libraries will be constructed in preparation of sequencing. For a more thorough description of the protocols used for generating viromes, see Chapter 3 in this volume.
There are currently four options when selecting a sequencing platform for metagenomic studies including di-deoxy sequencing (Sanger), pyrosequencing (454-Roche), SOLiD™ (Applied Biosystems) and Illumina® (formerly known as Solexa). Each technology has pros and cons with respect to sequencing performance including overall cost, read length, error rates, and total capacity (see Chapter 18, Vol. I). To date, only Sanger and 454 pyrosequencing have been utilized in viral metagenomic studies. Sanger sequencing, the only option available when the first viral metagenomic study was undertaken [Breitbart et al., 2002], still affords the longest read lengths of all available sequencing technologies [Wommack et al., 2008]. However, the sheer volume of data, increasing read lengths, and cost advantage afforded by pyrosequencing has resulted in a sharp decline in Sanger-based metagenomic projects in the past several years.
2.3 Environmental Studies
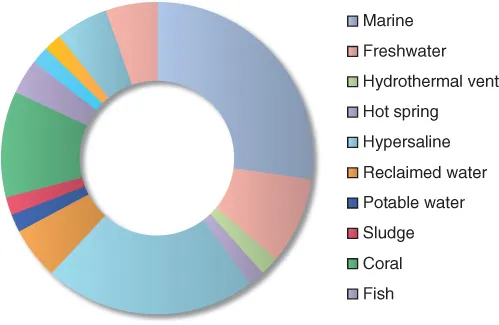
The diversity of natural ecosystems on our planet presents unparalleled opportunities for viral metagenomic studies, and a tremendous amount of data on virus communities inhabiting a multitude of environments has been produced over a relatively short time span. The majority of viral metagenomic studies have focused on dsDNA-containing viruses, although targeted studies of viruses with alternate nucleic acid types are increasing [Culley et al., 2006; Ng et al., 2009a,b; Andrews-Pfannkoch et al., 2010]. Due to the extensive nature of environmental viral metagenomic studies, it's prohibitive to discuss them all in detail. Therefore, this part of the chapter will highlight the significant observations generated from studies conducted on samples collected from aquatic, terrestrial, and extreme ecosystems. Figure 2.1 shows the distribution of environments from which viral metagenomes have been produced. Most studies have focused on the marine environment, although numerous hypersaline viromes have also been created. A viral metagenome has even been generated from a vineyard, effectively establishing a connection between the disciplines of viral ecology, plant pathology, and oenology [Coetzee et al., 2010].
Despite the disparate physical and chemical factors that characterize the environments shown in Figure 2.1, the resultant viromes generally share the following three characteristics: (1) a high incidence of unknown sequences, (2) a high level of genotypic diversity, and (3) evidence of functional and metabolic plasticity. The propensity of unknown sequences, or sequences that share no significant similarity to other sequences in public databases, suggests that environmental viruses are the most uncharacterized and genetically novel biological components on our planet. The high number of estimated virus genotypes in several environments undoubtedly contributes to the novelty of viral metagenomic data as well as substantial evolutionary divergence from viruses within public databases (see Chapter 4 in this Volume for additional information). Functional profiling of environmental metagenomes has perhaps revealed the most intriguing observations with respect to virus–host dynamics, adaptation, and evolution. A multitude of studies have now demonstrated that the adoption of environmentally relevant host genes by viruses, predominantly phage, is a common occurrence (see Rohwer and Thurber (2009) for a review of this topic; also see Dinsdale et al. [20...