In the words of the famous developmental biologist, South African–born Lewis Wolpert, who worked on developmental biology in the United Kingdom, “not birth, marriage or death, but gastrulation which is truly the most important time in your life” [1,2,3]. The cell differentiation from the stem cell gives it an identity with a specific, dedicated function. This process of cell fate determination (CFD) is initiated by the differentiation of embryonic stem cells (ESC) into distinct lineages giving an identity to the differentiated cell with unique structural and functional features. This process of decision making of cellular processes is the fundamental key to life. Cell fate (CF) choice involves intricate functioning of complicated networks, protein–protein crosstalk, and signaling reaction with the help of signaling molecules.
Thus to understand the molecular functions of cellular processes like cell differentiation, cell fate choices after DNA damage, or their differentiation into different cells from the progenitor stem cells, understanding of genetic switches, signaling pathways, and involvement of transcription factors (TFs) is important and at the same time profoundly complex. The adult stem cells located in the tissues are also thought to differentiate into specific lineages and lead to replacement of differentiated cells within regenerating tissues, like lungs, skin, blood etc.
Thus although it was thought to be a phenomenon associated with development, cell fate determination is now thought to be a phenomenon associated with the integral part of the biology of a cell. This also forms a major domain in controlling the pathways so that disease could be prevented and development may be controlled.
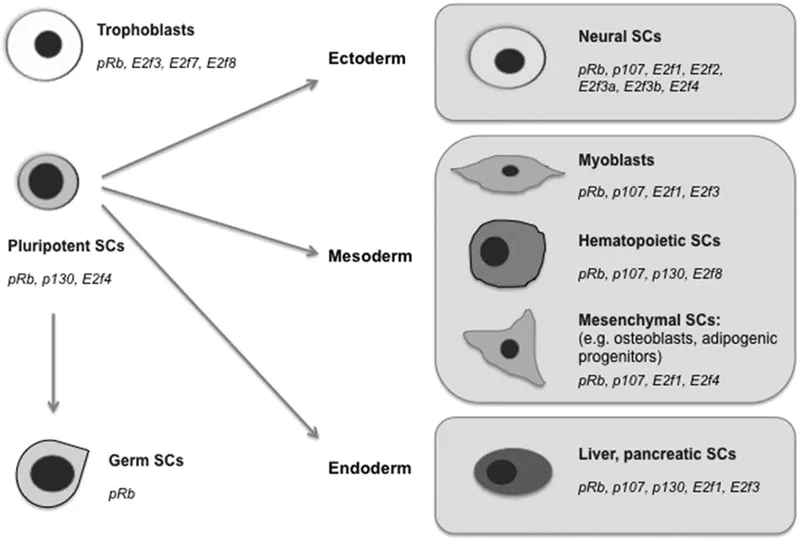
The decision on the determination of a cell’s fate regulates the behavior, morphology, migration pattern, proliferation, and function of the differentiated cell. The fate of stem cells is regulated by different factors (Figure 1.1).
Each type of lineage is committed to a special function. Like the cells in the islets of langerhans of the pancreas consisting of alpha and beta cells that produce glucagon and insulin respectively and form the endocrine part of the pancreas, the neurons with their specialized structures are committed to transmit impulses and the red blood cells (RBC) are committed to carry oxygen (O2) to the cells and remove carbon dioxide (CO2) from the cells. Likewise the immune cells are committed to confer immunity. Each differentiated cell is designed with specific cellular properties, like shape and size, that regulates its function.
For a progenitor stem cell differentiating into a fixed type of cell lineage, cell fate decisions are thought to be manifested by asymmetric cell divisions [4] while the surrounding microenvironment and niche have also been shown to signal their differentiation [5,6].
This process of differentiation involves the activity of specific transcription factors (TFs) dictating a particular lineage and downstream signaling networks working in unison to lead to the generation of a specific cell with a distinct identity. However, there are more questions than answers in this field of importance. Although cell fate determination is such an important event, the intricate biology regulating the specific morphology and function of a differentiated cell is not well understood. Two major domains require further research as to the factors and processes controlling a cell’s pathway towards its signaling network for differentiation. Computational approaches of digital imaging are helping us to understand this phenomenon. The path that leads to a cell’s identity, including its structure, function, and behavior, remains less studied, and attempts are being made to understand through recent developments in genomics (Figure 1.2).
High-resolution live cell imaging has enabled us to trace embryonic development in single cells, thereby generating complete cell lineage fate maps [7,8] with characterization of the single cell with its volume, shape, migration rates, trajectories, extensions, and contacts within a developing embryo [9,10], and indicative of the cell’s fate and gene expression map is helping us to understand the regulatory network involved in the process.
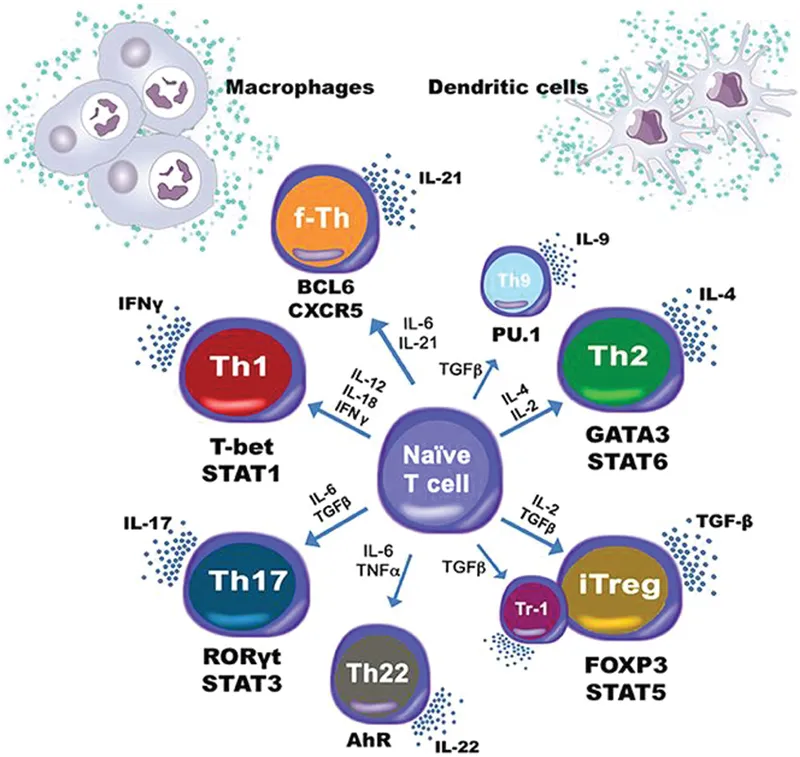
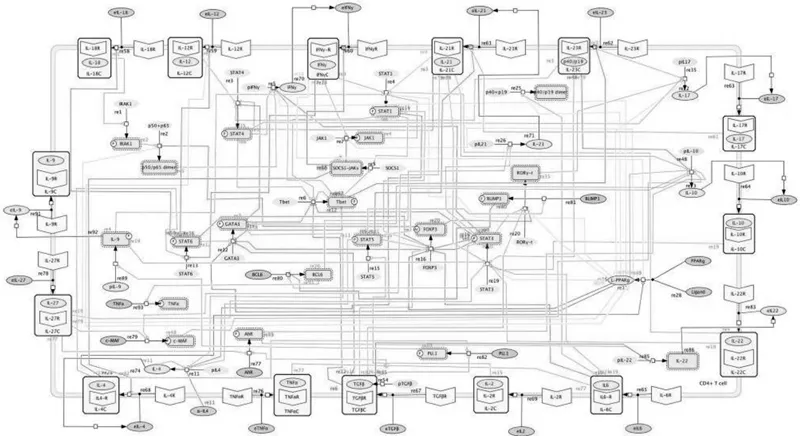
The immune system includes different cells with diverse signaling molecules and cascades. The coordination between the pro-inflammatory and regulatory cells enables tissue homeostasis, and CD4+ T cells play major role in this process. CD4+ T cell differentiation involves signaling mediated by cytokines and their receptors, adaptor molecules, and transcription factors (TFs) that help delineate cell fate and function. T cell subsets include Th1, Th2, Th17, Th9, Th22, follicular T helper cells (Tfh), induced regulatory T cells (iTreg) and type 1 regulatory T cells (Tr1) with diverse functions induced by different cytokines released by the dendritic cells and macrophages. Computational modeling is enabling better analysis of the complicated interplay of different molecules in CD4+ T cell differentiation (Figure 1.3).
1.3 COMPUTATION AND STEM CELL BIOLOGY AND CELL FATE
Over the last fifty years advances in computation have immensely furthered our understanding of life processes. Developmental biology with regard to the origin of a committed cell from a stem cell is one major domain being extensively studied today. Alan Turing wrote a computer program that for the first time could model pattern formation from morphogen concentration in an in silico embryo. Since then, computational technology with its improved application-based methodology has helped in the understanding of cell fate. Fluorescent in situ hybridization (FISH) studies have enabled generation of images [12,13] thereby generating an in silico embryo, on which in silico studies can be performed by overlaying the gene expression pattern [14]. With the generation of high throughput data from omics approach, including information on the TFs and chromatin profile, gene expression is being used in construction of GRNs of a cell in its transition and differentiated form [15,16,17] by generation of stem cell (SC) models.
Recent developments from studies from next generation sequencing (NGS [18,19]) and its application in single cell transcriptomics (SCT [20]) hold great promise in understanding the biology of a single cell and its role in development [21], and more research is still underway. Some of the methods that have been applied include OMIC-based analytical tools like the hierarchical clustering (HCL) that enables hierarchy building of clusters through a matrix-based approach by joining the closest pair of entities and considering the distance between all entities. Other methods include K-means clustering, which aims at grouping data into clusters and then detecting the inter- and intra-cluster distances. Principal component analysis (PCA) also finds application here. Differential analysis is another method that aims to identify differentially expressed genes by distinct groups.
Enrichment analysis (EA) uses the programme of gene set analysis (GSA) that determines the expression of predefined sets of genes and their clustering around the top or bottom. Mutation calling is another method used to identify the genetic differences between a sample with a reference. Peak comparison involves identification of NGS-enriched genomic loci generated from ChIP-seq or DNase-seq. Application of Bayes theorem (BT) called the naive Bayes classifiers (NBC) and random forest (RF) enabled analysis of data to understand gene expression between the stem cell and the differentiated cell. NBC is a probabilistic model of machine learning used for classification, based on Bayes theorem (BT) assuming strong ...