
This is a test
- 300 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Book details
Book preview
Table of contents
Citations
About This Book
This book assembles a comprehensive collection of plant virus electron micrographs of good quality, offers a consistent treatment, and backs the visual data with a consistent and comprehensive text. Although this book is primarily about the structure of virus particles and infected cells, the results of biochemical experiments are referred too when relevant, so that the virus particles described appear as part of a replicating complex. Similarly, infected cells are portrayed as active rather than static structures.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Atlas Of Plant Viruses by Robert G. Francki R.I.B; Milne in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Botany. We have over one million books available in our catalogue for you to explore.
Information
Chapter 1
COMOVIRUS GROUP
Comoviruses derive their name from the type member, cowpea mosaic virus (CPMV).1Each Comovirus appears to be related to at least one other group member, the group forming a network of serologically related viruses.2 The viruses have small isometric particles about 30 nm in diameter built from two species of polypeptides of MT, about 37 × 103 and 23 × 103 The particles sediment as three components which have identical protein coats but vary in their RNA content. They encapsidate two species of RNA of MT about 2.4 × 106 and 1. 4 × 106, which together constitute the viral genome.
Each Comovirus is transmitted to a narrow range of plants and most have been shown to be vectored by leaf-feeding beetles. Seed transmission usually occurs at a low but significant level.3 Comoviruses can be transmitted by mechanical inoculation. They all induce characteristic cytopathological effects in the cytoplasm of infected cells.2–4
I. Members of the Group and Their Relationships
CPMV, by far the best known member of the group, has been used extensively for studies on particle structure, mode of replication, and cytopathology. Most of the remaining Comoviruses have been studied rather superficially. However, all the available data, including those establishing extensive serological relationships among the members, make it fairly safe to assume that Comoviruses are all very similar.2-3-5 Nevertheless, the serological data are not sufficiently precise to draw clear conclusions about the degrees of relationship among the members. In many cases it is difficult to decide whether some of the described viruses merit separate names or should be considered as strains of the same virus. This has already introduced some confusion in the literature; for example, the virus now known as cowpea severe mosaic virus (CPSMV) was originally referred to as the severe strain of CPMV. There are now good reasons for considering the two viruses as distinct on the basis of differences in host range, symptoms produced, antigenic properties, nucleotide sequences, and their inability to produce viable pseudorecombinants.6-9 There is also confusion about the identity of viruses such as bean curly dwarf mosaic virus (BCDMV).10 Although the authors concluded that BCDMV was sufficiently similar to quail pea mosaic virus (QPMV) to be considered as a strain of that virus, they referred to each isolate by a distinct name.10
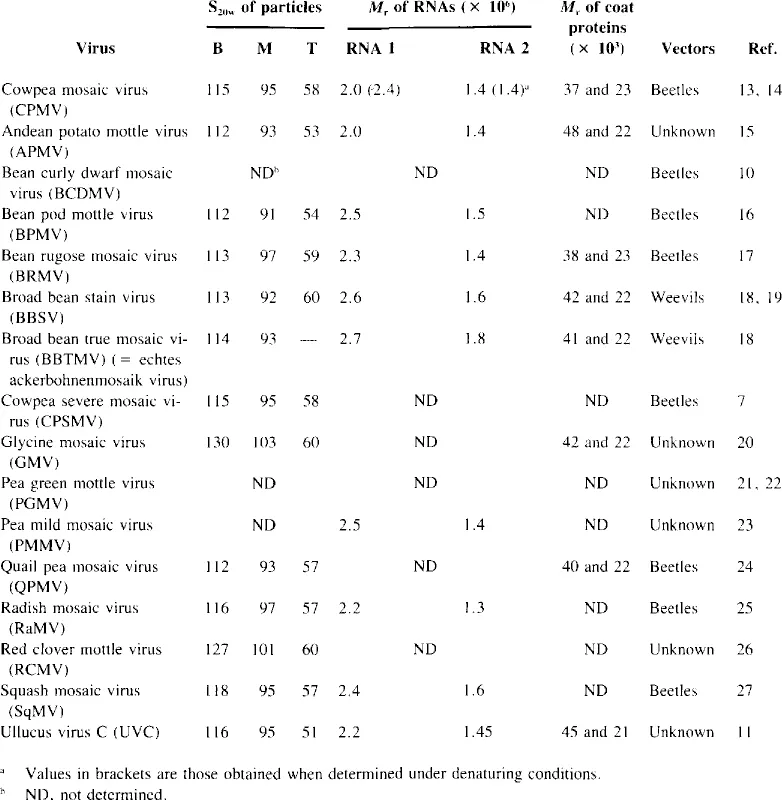
All but four Comoviruses infect predominantly leguminous plants. Radish mosaic virus (RaMV), squash mosaic virus (SqMV), andean potato mottle virus (APMV), and Ullucus virus C (UVC) usually infect plants within the families Cruciferae, Cucurbitaceae, Solan- aceae, and Basellaceae, respectively.2,11 However, from the limited serological data available, one cannot conclude whether these host range differences are reflected in the antigenic properties of the viruses.2,3 Much more work will be required to establish a satisfactory taxonomy of the group. This could be achieved with more extensive serological studies using uniform techniques throughout. Much useful taxonomic information might also be obtained by comparing nucleotide sequence homologies between the various Comoviruses and their strains. The ability to form viable pseudorecombinants between the isolates may also prove to be informative.12 For the present, a tentative list of Comoviruses is presented in Table 1. However, as more information becomes available, some of the listed viruses may prove to be more appropriately considered as strains.
Although broad bean wilt virus (BBWV) has many chemical and physical properties in common with the Comoviruses,28,29 its taxonomic position remains enigmatic. It has been assigned as a possible member of the Comovirus group,1 but does not appear to be serologically related to any of the Comoviruses and is unlike them in being transmitted by aphids.
Table 1
MEMBERS OF THE COMOVIRUS GROUP AND SOME OF THEIR PROPERTIES
MEMBERS OF THE COMOVIRUS GROUP AND SOME OF THEIR PROPERTIES
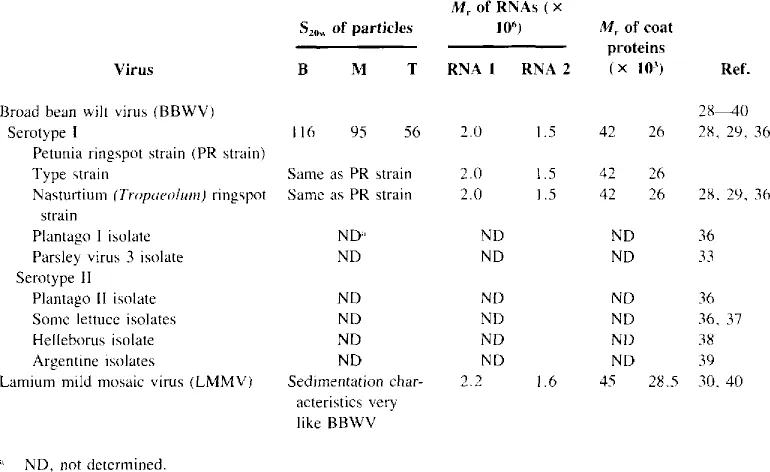
It has been suggested that BBWV and the serologically distantly related lamium mild mosaic virus (LMMV) (Table 2) should be considered as members of a new plant virus group.29,30
II. Virus Structure and Composition
A. Particle Structure
Electron micrographs of Comovirus particles (Figure 1) negatively stained with uranyl acetate and those of BBWV stained with potassium phosphotungstate (Figure 2) show that they are polyhedra about 30 nm in diameter. Crowther and colleagues41 reported the particle diameter of CPMV to be only 20 to 24 nm, and estimates of the diameters of other Com- oviruses by other workers range from 24 to 30 nm. No doubt some of this variation is due to differences in preparing the particles for electron microscopy and errors in the calibration of the microscopes used. When stained in uranyl acetate and compared directly to virus particles in a number of different taxa, broad bean stain virus (BBSV) and broad bean true mosaic virus (BBTMV) particles were found to be 30 nm in diameter.42
Table 2
SOME PROPERTIES OF BROAD BEAN WILT AND LAMIUM MILD MOSAIC
SOME PROPERTIES OF BROAD BEAN WILT AND LAMIUM MILD MOSAIC
Preparations of Comoviruses as well as those of BBWV and LMMV usually sediment as three components referred to as bottom (B), middle (M), and top (T) components, respectively (Table 1). All the particles have indistinguishable protein shells, but the T component is devoid of RNA, whereas the B and M components contain one molecule each of RNA 1 and RNA 2, respectively.2
The Comovirus protein shell is built from equimolar amounts of two distinct proteins of Mr about 37 × 103 and 23 × 103 (37K and 23K proteins, respectively).13 Early studies on CPMV structure were complicated by the detection of two electrophoretic components in most virus preparations. However, it was shown that this was caused by an in situ degradation of the smaller coat protein subunit by proteolytic cleavage in the host cells and after isolation of the virus.43 The cleavage involves the loss of a peptide of Mr about 2.5 × 103 From the size of the two intact proteins and the molecular weight of the CPMV protein shell, it has been calculated that each particle consists of 60 molecules of each of the two proteins.13,44Three-dimensional image analysis from electron micrographs of CPMV revealed that the particles have 5:3:2 symmetry. A model has been proposed for the CPMV particle consisting of 12 pentamers of the 37K protein at the fivefold positions and 20 trimers of the 23K protein at the threefold positions (Figure 3).45
B. The Viral Genome
The CPMV genome is bipartite.2,45 It is composed of two single-stranded, positive-sense RNA molecules, both of whose complete sequences have been determined.46,47 RNA 1 from B component capsids consists of 5889 nucleotides,47 and RNA 2 from the M component of 3481 nucleotides; that is, excluding their poly A tails of 150 to 200 residues at their 3’ termini.48 Although the sequences of the two RNAs are quite distinct, they are similar adjacent to these tails.49 At their 5’ termini, both RNAs have a covalently ...
Table of contents
- Cover
- Title Page
- Copyright Page
- Preface
- Authors
- Acknowledgments
- Table of Contents
- Chapter 1 Comovirus Group
- Chapter 2 Nepovirus Group
- Chapter 3 Pea Enation Mosaic Virus Group
- Chapter 4 Dianthovirus Group
- Chapter 5 Cucumovirus Group
- Chapter 6 Bromovirus Group
- Chapter 7 Ilarvirus Group
- Chapter 8 Alfalfa Mosaic Virus Group
- Chapter 9 Tobamovirus Group
- Chapter 10 Hordeivirus Group
- Chapter 11 Tobravirus Group
- Chapter 12 Potexvirus Group
- Chapter 13 Carlavirus Group
- Chapter 14 Potyvirus Group
- Chapter 15 Closterovirus Group
- Chapter 16 Some Unclassified Viruses
- Virus Abbreviations and Taxonomic Groups
- Index