
This is a test
- 370 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Modeling the 3D Conformation of Genomes
Book details
Book preview
Table of contents
Citations
About This Book
This book provides a timely summary of physical modeling approaches applied to biological datasets that describe conformational properties of chromosomes in the cell nucleus. Chapters explain how to convert raw experimental data into 3D conformations, and how to use models to better understand biophysical mechanisms that control chromosome conformation. The coverage ranges from introductory chapters to modeling aspects related to polymer physics, and data-driven models for genomic domains, the entire human genome, epigenome folding, chromosome structure and dynamics, and predicting 3D genome structure.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Modeling the 3D Conformation of Genomes by Guido Tiana, Luca Giorgetti in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.
Information
1
Chromosome Folding: Contributions of Chromosome Conformation Capture and Polymer Physics
Job Dekker
1.1 Introduction
1.2 Chromosome Conformation Capture
1.33 C Variants to Obtain Genome-Scale and High-Resolution Chromatin Interaction Matrices
1.4 Insights Obtained From Chromosome Interaction Data
1.5 Dynamics and Cell-to-Cell Variation in Chromatin Interactions
1.6 Polymer Models for Chromosome Folding
1.7 Mechanisms of Chromosome Folding and Nuclear Organization
1.8 Future Perspective
Acknowledgments
References
1.1 Introduction
It is now hard to imagine that there was a time when there was a debate about the extent to which the genome is organized in any specific way inside the interphase cell nucleus or nucleoid. One reason for this debate is that in some experiments chromatin can appear highly structured, e.g., forming hierarchies of increasingly folded and thicker fibers (1), while in other experiments very little structure is detected in what appears to be an ocean of nucleosomes (2). Further, fluorescence in situ hybridization experiments to localize specific loci reveals tremendous cell-to-cell variability in sub-nuclear position and distance between any pair of loci. However, these experiments also show some clear trends. For instance, chromosomes occupy their own territories (3) with some limited intermingling at their borders (4). The positions of chromosomes can vary greatly between individual cells, but some chromosomes and chromosomal domains tend to be more often at the periphery of the nucleus while others are more often near the center of the nucleus (5, 6). Further, in general, euchromatic loci have been observed to associate with other euchromatic loci, and heterochromatic loci to interact with other heterochromatic loci (3, 7). Combined, these and many other findings suggest that nuclear organization is variable and stochastic on the one hand, while also guided by some common principles and that this relates to whether chromatin is (transcriptionally) active or inactive (8). Further, chromosomes are obviously organized in some defined manner during mitosis when the classic rod-shaped structures are observed. However, even in that case, how chromatin is folded during mitosis (and meiosis) has been debated for many years.
Over the last two decades, there has been enormous progress in our understanding and appreciation of the spatial organization of genomes and how it mediates or modulates the many functions of chromosomes such as the regulation of gene expression, the repair and replication of DNA, and chromosome condensation and transmission to daughter cells. This has been driven by improved imaging techniques, including super-resolution and live cell approaches but most particularly by two developments that, as outlined in the following, are significantly interlinked: First, the development of molecular genomic approaches for mapping the structure of chromosomes, mainly based on chromosome conformation capture (3C) technology; and, second, the development and application of insights from the field of polymer physics to the problem of chromosome folding.
The debate has now moved to mechanistic interpretations of structural features observed with different technologies, their dynamical properties, how variable these structures are between otherwise identical cells, and how chromosome architecture instructs or influences any of the genome’s functions. Here I outline the conception of 3C, it’s development over the last two decades, and how it has stimulated interactions between cell biologists, molecular biologists, (polymer) physicists, mathematicians, and computational biologists. This rich interdisciplinary interface is now producing remarkable new insights into the chromosome folding problem, discussed in this book.
1.2 Chromosome Conformation Capture
The introduction of chromosome conformation capture in 2002 (9) has revolutionized the study of the spatial organization of genomes by allowing mapping of three-dimensional chromosome structure at increasingly high resolution directly to sequence and at the scale of complete genomes. The key new concept that motivated the subsequent development of 3C was the idea that when a matrix of many or all interaction frequencies between and among loci located along a chromosome could be measured, the three-dimensional organization of that chromosome could be inferred. 3C is used to detect the frequency of interaction of any pair of genomic loci, and when combined with deep DNA sequencing can generate genome-wide all-by-all interaction frequency matrices (Figure 1.1). Such matrices have now been shown to indeed allow the derivation of models of the spatial organization of chromosomes and even help gain insights into the dynamics and mechanisms of their folding.
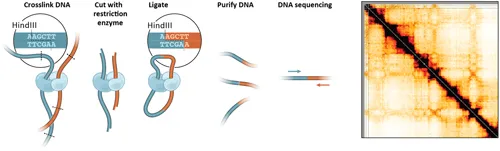
Figure 1.1 Schematic outline of the key steps of 3C-based assays. Left: Chromatin is crosslinked, and then digested and religated. (adapted from (23)). Ligation products are then sequenced. 3C variants such as 4C, 5C, and Capture C differ in how ligation products are detected, or include steps to label digested ends (Hi-C), or include a step to selectively purify fragments bound by specific proteins (ChIA-PET, HiChIP). Right: 3C-based assays are used to obtain matrices of interaction frequencies that can be depicted as heatmaps. This example shows an interaction matrix for human chromosome 21 in HeLa S3 cells. The color intensity reflects relative interaction frequencies.
In 3C cells are fixed with formaldehyde (Figure 1.1). This essentially freezes the spatial arrangement of chromosomes in place. The spatial proximity between loci is then determined by fragmenting the chromatin with a restriction enzyme, while the chromatin remains frozen in place. Spatially proximal DNA ends are subsequently re-ligated and DNA is purified. This assay produces a large collection of unique DNA ligation products that each represents a spatial co-location event in one of the cells in the population. Given that DNA molecules are easily identified and characterized by PCR or DNA sequencing 3C reduces the difficult problem of determining relative spatial positions of loci inside cells to the much simpler process of DNA sequence analysis.
3C combines a number of molecular steps that had previously been used separately, e.g., proximity ligation had been used for many years to detect protein-induced bending and looping of DNA in vitro and in vivo (10–12). The innovation of 3C lies in the fact that it was designed to be unbiased and able to detect any spatial proximity, even when not mediated by a specific factor, so that it can detect all the spatial proximities irrespective of the specific mechanisms that brought the loci near each other. The concept on which it is based is also innovative, asserting that dense matrices of interaction frequencies reveal the principles of chromosome folding.
Initially, PCR was used to read and quantify specific ligation product formation events, e.g., to determine whether specific loci would interact with each other more frequently than expected. In the original 3C paper, previously known specific interactions between yeast centromeres, between telomeres, and between homologous chromosomes could be detected, thereby validating the approach (9). Further, the first (albeit sparse) matrix of interaction frequencies for a complete chromosome was presented and this matrix was used, through polymer physics theory and mathematical optimization to infer the population average folding of this chromosome. Of note, this very early work already employed polymer models for analysis of 3C data. Polymer theory and model building to interpret matrices of chromatin interaction frequencies has become a field of intense study and is now contributing to fundamental new insights into how chromosomes fold, as is outlined throughout the chapters in this book.
3C was rapidly adopted and in several landmark publications it helped demonstrate that enhancers could loop to their target genes, e.g., in the beta-globin and alpha-globin loci, in a tissue-specific manner and it was found that such specific looping interactions depend on specific transcription factors (13–19). Although such locus-specific studies to map interactions between and among specific elements in loci of interest continue to this day, further innovations in 3C technology now enable genome-wide and unbiased mapping of chromatin interaction matrices.
1.33 C Variants to Obtain Genome-Scale and High-Resolution Chromatin Interaction Matrices
The size of genomes, and thus the number of possible chromatin interactions, makes 3C analysis using PCR impractical. Since the initial development of 3C, many experimental adaptations have been introduced that allow the mapping of chromatin interactions at a genome-wide scale, with increased resolution, in cell populations and in single cells. All these variants follow the basic 3C protocol of crosslinking chromatin, DNA fragmentation and relegation of DNA ends that are in close spatial proximity. They differ in the method for ligation product detection.
The first two 3C variants, 4...
Table of contents
- Cover
- Half-Title
- Half-Title
- Title
- Copyright
- Contents
- Preface
- Editor
- Contributors
- 1 Chromosome Folding: Contributions of Chromosome Conformation Capture and Polymer Physics
- PART 1 FIRST-PRINCIPLES MODELS
- PART 2 DATA-DRIVEN MODELS
- 15 Index