This section deals with the principal structural characteristics of DNA. We should point out at once that atomic structure is beyond the scope of this book. We are going to look at structure primarily as it relates to the analysis of circular DNA and its properties. Therefore, many important issues will be overlooked or barely touched upon. For a more comprehensive overview of DNA structure, the reader is referred to the monographs and surveys cited in this chapter.
A. Single-Stranded DNA
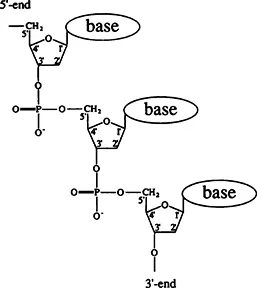
Single-Stranded DNA consists of a sugar-phosphate polymeric backbone and lateral nitrous bases attached to it. The chemical structure of single-stranded DNA is presented in Figure 1. The backbone’s repetitive unit (link) includes six skeletal bonds. In each chain link one of four bases, adenine (A), guanine (G), cytosine (C), or thymine (T), is attached to the C1′ atom of the sugar ring. These bases (see Figure 2) belong to two different chemical groups; adenine and guanine are purine bases, while cytosine and thymine are pyrimidine bases. The base sequence, unlike the sugar-phosphate chain, is irregular, for it contains the coded genetic text carried by the DNA molecule. As can be seen in Figure 1, the backbone has no symmetry relative to the movement along it one way or the other; the structure of the backbone defines the chain direction. One end of the chain is called the 3′ end (lower end in Figure 1) and the other is called the 5′ end (upper end in Figure 1). These appellations have to do with the numbers of carbon atoms in deoxyribose. To define the chemical structure of a single-stranded DNA, it is sufficient to specify the base sequence. By a generally accepted convention, the sequence is always written from the 5′ end towards the 3′ end. Rotation may occur around each single bond of the sugar-phosphate backbone. Naturally, this rotation is not free because a number of rotation angles (called dihedral angles) entail too close contacts between atoms adjacent to the principal chain. Still, the permissible dihedral angle ranges are large enough. (For more detailed and rigorous presentation of this question, the reader is referred to the Cantor and Schimmel textbook.1) Therefore, the sugar-phosphate backbone is highly flexible; even one repeating unit can assume a very large number of conformations. Without causing any major tensions in the structure, one can adapt the sugar-phosphate backbone to a variety of conformational requirements. This property is responsible for the polymorphism of helical structures that one observes in double-stranded DNA. Note that the size of a repeating unit in a conformation stretched to the limit is about 0.7 nm.
FIGURE 1. Chemical structure of single-stranded DNA.
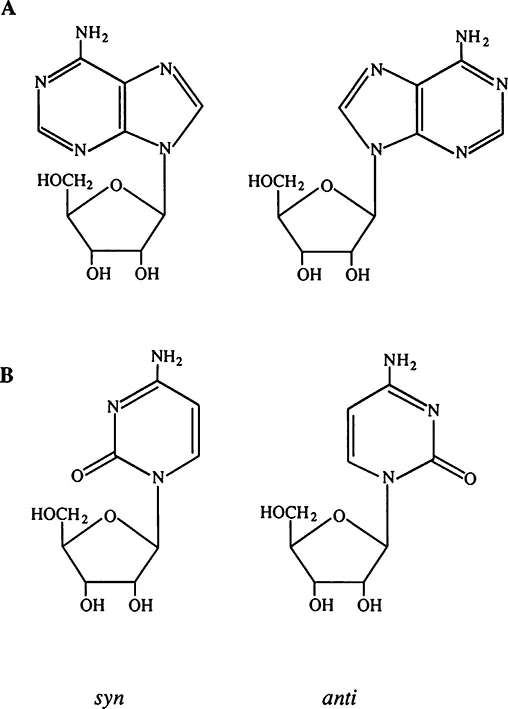
Apart from rotation around backbone bonds, there is also the very important rotation around the so-called glycosidic bond between the sugar and base. Permissible rotation angles around the glycosidic bond largely fall within two ranges with almost 180° in between. The nucleotide link conformations corresponding to these two ranges are designated as syn and anti. Figure 3 shows the syn and anti conformations of purine and pyrimidine. A large body of calculations and experimental data obtained with monomers demonstrates that the two conformations are roughly equivalent energy-wise for purines, whereas the anti conformation is the referred one for pyrimidines.1
Finally, another degree of freedom of the polynucleotide chain is associated with the conformation of deoxyribose. This ring’s conformation is not flat. As a rule, four atoms lie on a plane, while the fifth one, either C2′ or C3′, is situated above or below the plane. Depending on whether or not this atom is shifted off the plane in the same direction as C5′, the conformation is denoted as endo or exo, respectively. Normally, C2′-endo and C3′-endo conformations occur in regular nucleic acid structures.
The sugar-phosphate chain carries a single negative charge per phosphate group. This has an appreciable effect on the conformational properties of DNA. Various consequences of DNA’s polyelectrolyte nature will be discussed throughout this book.
B. The Double Helix
Single-stranded DNA is a relatively rare thing under natural circumstances. One can say that DNA’s regular form is a double helix consisting of two antiparallel single strands. The discovery of the double helix by Watson and Crick in 1953 is regarded as the birthdate of molecular biology. More than the structure of the genetic information carrier was discovered at that time. Even a fleeting glance at this structure makes it clear that it contains the mechanism of a fundamental genetic process, viz., the duplication of genetic information. No biological structure decoded before or after that momentous discovery proved nearly so revealing of the molecular functioning mechanisms.
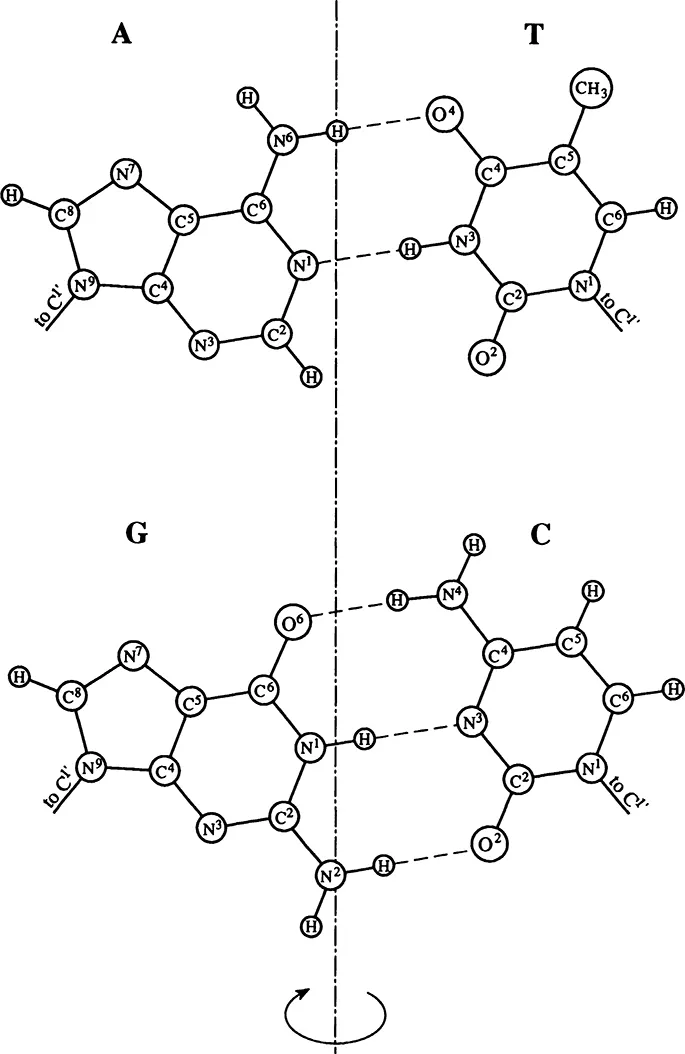
FIGURE 2. Complementary base pairs. On turning around the pseudosymmetry axis that passes between pairs, the conds N9-C1′ and N1-C1′ trade places.
FIGURE 3. Anti and syn conformations of purine (A) and pyrimidine (B).
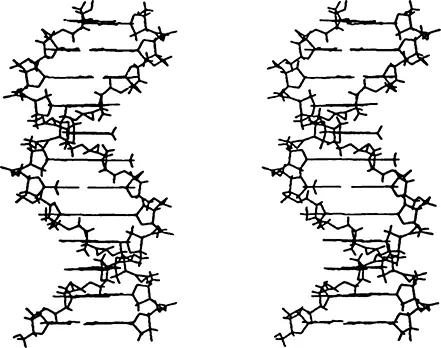
A three-dimensional model of the double helix (B-form) is shown in Figure 4.3 The bases in this structure are inside the helix, while sugar-phosphate chains are outside, running in opposite directions. Thus, the double helix, unlike a single strand, does not have a structurally defined direction. The bases, one from each strand, form pairs linked by hydrogen bonds. Significantly, only two types of pairs, AT and GC, are possible in a regular double helix. These pairs have the same geometry with regard to their bonds to the sugars. Besides, their symmetry lane passes through the helix axis (see Figure 2). Therefore, they are readily inscribed in the regular double helix structure. These pairs are called Watson-Crick, or complementary, pairs because the two bases complement each other in forming the characteristic pair geometry. The requirement that the bases within a pair be complementary is the reason why the sequence of one strand fully defines the sequence of the other strand.
FIGURE 4. Skeleton model of B-DNA double helix. (From Schlick, T., et al., Theoretical Biochemistry and Molecular Physics, Volume 1: DNA, Adenine Press, Schenectady, NY, 1980. With permission.).
The base pairs are situated inside the helix and cling tightly to each other, forming a pile. That is why they have almost no contact with water molecules...