
This is a test
- 233 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Precision Haematological Cancer Medicine
Book details
Book preview
Table of contents
Citations
About This Book
Many experts now consider genetic evaluation to be pivotal for the optimal diagnosis, classification, risk stratification, and therapeutic decision-making for persons diagnosed with blood cancer. This new text specifically focuses on the genetic alterations essential for establishing diagnosis and assesses how they might impact the precision oncology standard of care. Providing an authoritative review of the state of the art, this is essential reading for physicians, hematologists, and oncologists for optimal management of individual patients.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Precision Haematological Cancer Medicine by Tariq Mughal in PDF and/or ePUB format, as well as other popular books in Medicine & Public Health, Administration & Care. We have over one million books available in our catalogue for you to explore.
Information
1Precision Medicine and Future Therapeutics
According to Nicholas Culpepper in his 1652 book The English Physitian, ‘a man may preserve his Body in Health; or cure himself, being sick, for three pence charge, with such things only as grow in England’. Prescient words, in some respects – it’s still all about giving the right patient the right treatment, at the right dose and time, but today it’s called precision medicine. The term has been introduced in cancer medicine to reflect the advances in our ability to characterize cancers at a molecular level and develop targeted treatments that might help counteract them. The term, however, is often imprecisely defined. It is used interchangeably with the terms personalized medicine, precision oncology, personalized oncology, genomic medicine and others. Collectively, none of these terms are precisely defined either. The word ‘precision’ implies exactitude and accuracy, whilst ‘personalized’ refers to adaptation in meeting an individual patient’s cancer requirements. Since most cancers tend to be complex, even when they exhibit a unique molecular signature, to be ‘precise’ we need to ‘personalize’ our approach. Personalized cancer medicine focuses on the optimal management of a patient and is driven by the presence of specific molecular characteristics and sometimes other distinguishing characteristics. Its principal objective is to improve the clinical outcomes of the individual patient. Arguably, the principles of precision medicine have been a cornerstone of medical practice since the earliest efforts to classify disease and prescribe specific treatment based on a diagnosis. What is new, however, is the astonishing pace of advances in genetics and the impressive diagnostic and treatment options to improve efficacy and cost-effectiveness. Pari passu, we recognize many challenges, not least those related to the cancer cell’s physiology and the cellular environment that may define the ecology of the cancer cells. We now stand at the threshold of multiple changes which, in turn, will determine how precision cancer medicine will define twenty-first-century cancer therapeutics.
Introduction
Over the past three decades, haematological malignancies have led the development of precision medicine, with the notable improvement in the understanding of the molecular pathways. This has been made possible by the increasing utility of ‘omics’, a holistic system comprising of genomics (genes), proteomics (proteins), epigenomics (modifications to chromatin affecting gene expression) and metabolomics, concomitantly with improved (and cheaper) technologies and computational bioinformatics. Haematological malignancies encompass a broad group of cancers, some of which originate from haematopoietic stem cells (HSCs), and others, such as some acute leukaemias, most lymphomas and plasma cell neoplasms originate from differentiated cells (Figure 1.1). The notion of HSCs acquiring a molecular aberration is of significant interest since, in general, these cells do not divide, and they are not prone to acquiring somatic mutations. Unsurprisingly, therefore, this genetic stability makes blood cancers rare, in comparison to the prevalence of solid tumours, such as lung, breast and colorectal cancers.
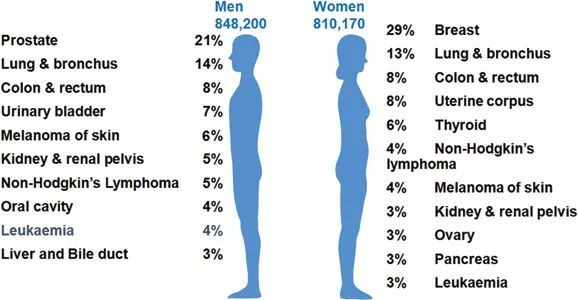
Figure 1.1Cancer statistics in the United States. (Data from the American Cancer Society; Siegel, RL et al., CA Cancer J Clin, 66:7–30, 2016.)
As a pedagogical device, haematological malignancies are divided into three broad categories, leukaemias, lymphomas and myelomas; however we know that the three categories have as many similarities as differences. The diversity of haematological malignancies reflects both the complexity of the blood system and the diversity of the molecular mechanisms that regulate haematopoiesis. It is of considerable interest that the diverse haematological malignancies manifest as disorders at different stages of haematopoietic differentiation. Leukaemias affect both haemopoietic stem cells and progenitor cells within myeloid and lymphocytic lineages (Figure 1.2). Lymphomas arise from lymphoid cells that, for example, in the case of T-cell lymphomas reflect blocks at multiple levels of T-cell differentiation from lymphoid precursors to T-follicular cells. Myelomas are a clonal disorder of plasma cells, which represent the final differentiation stage of B-cells. Leukaemias, lymphomas and myelomas encompass a large number of subtypes with a broad range of natural histories, ranging from those which remain indolent for long periods, to those that grow rapidly and can prove fatal very quickly if left untreated. The diagnosis of haematological malignancies relies on the integration of multiple diagnostic tools, involving morphology, flow cytometry, cytogenetics and molecular genetic analyses. And the utility of this in a systematic manner has been pivotal in order to provide the information necessary for optimal management of patients afflicted with haematological malignancies.
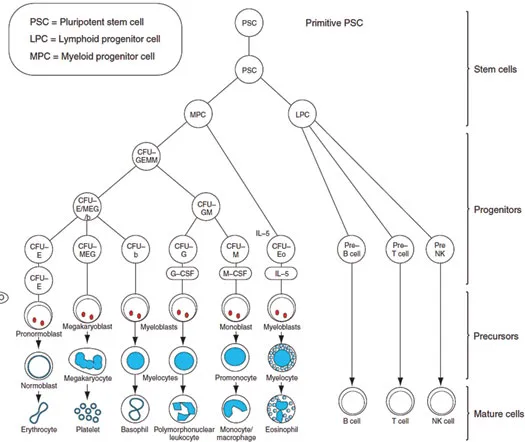
Figure 1.2A schematic representation of hemopoiesis. Abbreviations: CFU-GM, colony-forming units for granulocytes and monocytes; EPO, erythropoietin; NK, natural killer.
Leukaemias are a group of disorders characterized by the excessive accumulation of abnormal white cells in the bone marrow and peripheral blood. Lymphomas are a heterogeneous group of cancers that originate in lymphoid cells in lymph nodes or other lymphoid tissue. Some, such as Hodgkin lymphoma (HL) and certain subtypes of aggressive non-Hodgkin lymphomas (NHL), are well-characterized; the remainder constitute a rather motley collection of conditions, ranging from those with a very indolent natural history to those which are very aggressive and, unless treated promptly, rapidly fatal. The phrase ‘NHL’ was coined to cover this latter group of diseases and has considerable historical context. It is also intriguing that different leukaemias and lymphomas are prone to occur at different ages. Multiple myelomas, used interchangeably with simply ‘myelomas’ or ‘plasma cell neoplasms’, arises from the plasma cells in the bone marrow and is characterized by the production of a single species (monoclonal) of immunoglobulin molecules (a paraprotein; also called M-protein).
Though the first molecularly targeted cancer medicine was probably tamoxifen, an anti-oestrogen medication, it was really the introduction of two drugs in clinics in 1998, imatinib mesylate (Novartis Pharma) for the treatment of patients with BCR-ABL1 positive chronic myeloid leukaemia (CML), and trastuzumab (Genentech-Roche Pharma) for the treatment of patients with HER-2 amplified breast cancer, which are credited for ushering in the personalized cancer medicine era (Figure 1.3). Now the term ‘precision immunology’, used interchangeably with the term ‘targeted immunology’, has also been introduced to describe the use of immunotherapy based on our enhanced understanding in tumour immunology, and the immune system’s ability to recognize tumour-associated antigens and mediate a highly specific immune-based attack on the cancer cells.
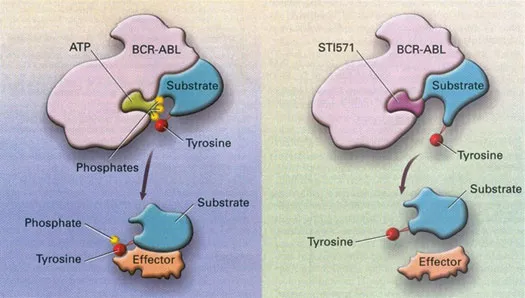
Figure 1.3Mechanism of action of imatinib mesylate. (Reproduced from Goldman, JM, and Mughal, TI, Postgraduate Haematology, 6th edition, Wiley, 2006. With permission.)
Genetics and the Genomics of Cancer
Life begins as a single cell (from the fertilized egg) that divides and whose progeny divides repeatedly according to a unique set of coded instructions present in their nuclei. There are about 1013 cells in the human body that are the product of 1016 cell divisions. The nucleus harbours about 30,000 genes. Each gene consists of deoxyribonucleic acid (DNA), and it is the DNA that determines exactly how our bodies are assembled and gives us our individual characteristics. Much of this has been learned over the past six decades, largely from progress in biomedical science, and more specifically in genetics and genomics, owing to an increasing understanding of the cellular and molecular mechanisms underlying the disease process. The work of James Watson and Francis Crick in Cambridge, and Maurice Wilkins and Rosalind Franklin in London, led to the discovery in 1953 that DNA in cells consisted of two complementary strands organized as a double helix. This and other related work paved the way for the Human Genome Mapping Project, a large, multinational effort that had amongst its goals determining the complete human genetic sequence, which began in 1990 and was completed in 2003, at an astonishing cost of $2.7 billion.
The terms ‘genetics’ and ‘genomics’ are often incorrectly used interchangeably. Genetics is the study of inherited traits or characteristics, whilst genomics is the study of the structure and composition of the material encoding these genetic instructions. The genetic information is coded by the sequence of four deoxyribonucleotides (adenine, thymine, cytosine and guanine) that comprise the ‘genetic alphabet’ within the DNA. Cellular DNA usually exists as a double stranded helix, in which one DNA strand is ‘zippered’ to the second fully complementary DNA strand. If each strand of DNA was to be fully stretched, it would measure about 1.8 m. In the cell, the DNA strands are wrapped around molecules called histones, which play a pivotal role in regulating expression of the genes. The human genome, defined as the complete set of genetic material present in a cell, spans 3 billion units of DNA, and in the dividing cell, the entire genome is arranged into bundles of strings, known as chromosomes, within the nucleus. The genome is organized into about 30,000 genes; each gene encodes a protein and includes regulatory elements that control synthesis of the protein. In order to understand the causes of cancer, we need to understand what actually goes wrong in our DNA that makes healthy tissue turn cancerous. Scientists have now uncovered most, but by no means all, genes that can cause cancers, should those genes mutate.
Genes have two clear-cut functions. The first, except for genes encoding functional RNAs, including ribosomal RNA (rRNA) and transfer RNA (tRNA), is to produce messenger ribonucleic acid (mRNA) which, in turn, produces proteins. The gene does this when it is ‘switched on’, for example, by a specific message to its DNA, and the cell that harbours it will respond by synthesizing a particular protein. The second function of genes is to replicate themselves precisely and thereby allow DNA replication. This is a complex process requiring the presence of many enzymes and other substances. The DNA determines exactly how our bodies are assembled, gives us our individual characteristics and carries the directions that a cell uses to perform a specific function. The completion of the Human Genome Mapping Project in 2003 sequenced the entire genome and established the DNA sequence of all known human genes. This knowledge, in turn, has created a dictionary of the human genome which we can interrogate through sophisticated bioinformatics systems. This genome dictionary has led to the firm integration of genomic sciences into clinical practice, not only for cancer patients, but rather all medical and surgical subspecialties. This progress has also led to the technology utilized to become significantly affordable and reliable. As the costs plummeted to less than $1000, initial low- and medium-throughput sequencing efforts, such as the Sanger DNA sequencing technology developed by Frederick Sanger in Cambridge (United Kingdom) in 1977, have paved the way towards parallel or next-generation sequencing (NGS) (Figure 1.4). Then in the mid-1990s, in the same city, Cambridge, Sir Shankar Balasubramanian, working with David Klenerman, invented the leading NGS technology, which is now central to all Illumina sequencing platforms.
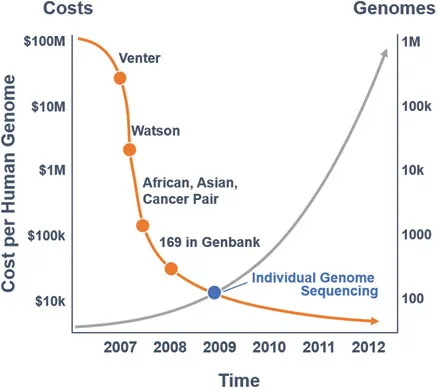
Figure 1.4The evolution of molecular testing. (Data from Wadman, M, Nature, 452(7189):788, 2008; https://www.genome.gov/27565109/the-cost-of-sequencing-a-human-genome/ [Accessed September 2017].)
Cancer Pathogenesis
Our interest in cancer pathogenesis has been richly rewarded since the 1980s, because of the impressive growth in understanding of cancer in general and the cancer genome in particular. The capacity to sequence cancer genomes accurately and precisely, has helped to garner a deeper understanding and comprehensively catalogue genomic alterations and novel oncogenic pathways in diverse cancers. Such efforts also underscore the conceptual role in cancer pathogenesis and therapeutic approaches of epigenetic alterations, alterations in the RNA processing and translation, engagement of the immune system and its relationship with the cellular environment, interplay with chronic inflammation and metabolic and hormonal milieu. Indeed, many of the advances highlight how little we understand the true complexity of the cancer ecosystem, a point that I return to in subsequent chapters.
All cells in the body appear to have three possible fates: they may proliferate to produce more cells, differentiate to carry out specialized functions or die at a predetermined time by a process termed apoptosis. The body requires an appropriate balance of cells undergoing each of these fates for normal function and survival. Normal cells usually die after 40–60 cycles of replication. In contrast, cancer arises when proliferation consistently and aberrantly exceeds apoptosis in a single (clonal) population of cells. Research into the mechanisms underlying the precise nature of how normal cell proliferation becomes abnormal and chaotic, and how these cells lose the ability to die at the prescribed time, suggests that this process has multiple stages, including pre-malignant changes, as first suggested by Isaac Berenblum in...
Table of contents
- Cover
- Half Title Page
- Title Page
- Copyright Page
- Contents
- Foreword
- Preface
- Acknowledgements
- Chapter 1 Precision Medicine and Future Therapeutics
- Chapter 2 Classifications and Risk Factors
- Chapter 3 Historical Milestones
- Chapter 4 Acute Myeloid Leukaemia
- Chapter 5 Acute Lymphoblastic Leukaemia
- Chapter 6 Chronic Myeloid Leukaemia
- Chapter 7 Myeloproliferative and Related Neoplasms
- Chapter 8 Myelodysplastic Syndromes and Myelodysplastic/Myeloproliferative Neoplasms
- Chapter 9 Leukaemias of Mature B- and T/NK-Cells
- Chapter 10 Hodgkin Lymphoma
- Chapter 11 Non-Hodgkin Lymphoma
- Chapter 12 Plasma Cell Neoplasms
- Chapter 13 Principal Treatment-Related Side Effects
- Chapter 14 Concluding Thoughts
- Bibliography
- Index