
This is a test
- 272 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Viral Regulatory Structures And Their Degeneracy
Book details
Book preview
Table of contents
Citations
About This Book
This book focuses on the nature, origins, and degeneracy (or redundancy) of viral regulatory elements and on the strategies that enable viruses to adapt to cells, examining experimental findings and models regarding HIV and HPV regulatory mechanisms.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Viral Regulatory Structures And Their Degeneracy by Gerald Myers in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.
Information
Howard Hughes Medical Institute and Department of Genetics, Duke University Medical Center, Durham, NC 27710; E-mail: [email protected]
Role and Mechanism of Action of the HIV-1 Rev Regulatory Protein
Reverse transcription of the retroviral RNA genome results in the formation of a double-stranded DNA proviral intermediate that is then integrated into the genome of the host cell. The resultant provirus is flanked by two long terminal repeats (LTRs). The 5′ LTR functions as the proviral transcriptional regulatory element while the 3′ LTR mediates efficient polyadenylation of the resultant transcripts (Figure 1). The large majority of retroviruses contain this single LTR promoter element, so that the primary transcript encoded by these retroviruses is identical to the viral RNA genome. The presence of only one promoter element, when combined with constraints on retroviral genome size imposed by the compact nature of retroviral virions, has forced retroviruses to rely primarily on posttranscriptional mechanisms to regulate, and facilitate, the expression of the various viral gene products. Of these mechanisms, the most important is clearly regulated alternative splicing.
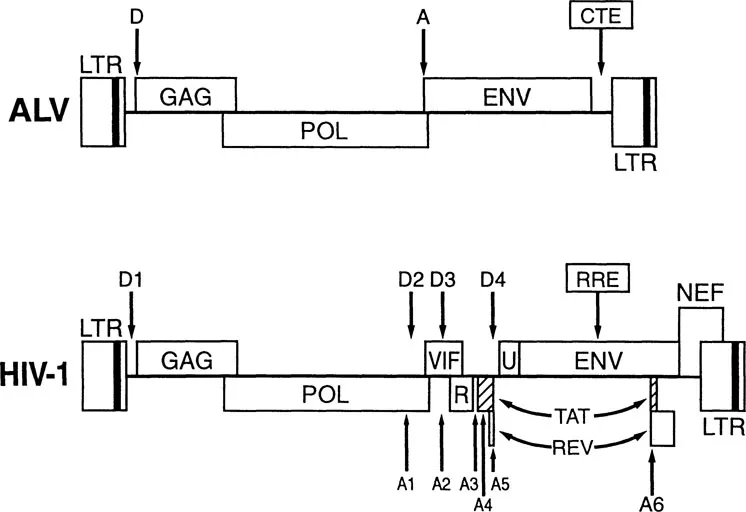
FIGURE 1 Genomic organization of ALV and HIV-1. Comparison of the genomic organization seen in ALV, a typical simple retrovirus, with the more complex HIV-1 genome. Viral genes are indicated, as are known major splice donor (D) and acceptor (A) sites. The localization of the Rev response element (RRE) and constitutive transport element (CTE) RNA targets are given. Abbreviations: LTR, long terminal repeat; R, Vpr; U, Vpu.
ALTERNATIVE SPLICING AND NUCLEAR RETENTION OF RETROVIRAL RNAs
Retroviruses do not encode any splicing factors, so that splicing of retroviral RNAs is controlled by the interaction of cellular splicing factors with cis-acting sequences in the viral transcript. In the case of the simple retrovirus avian leukemia virus (ALV), which encodes only a full length and a singly spliced transcript, an approximately 1:1 ratio between these mRNAs is maintained 1/M primarily as a result of a suboptimal splice acceptor site at the beginning of the viral env gene (Figure 1).13 Mutations of this 3′ splice site, which render its sequence closer to consensus, and, hence, improve splicing efficiency, result in an inhibition of viral replication and lead to the selection of viral revertants that, upon analysis, are found to again splice inefficiently.
The genome of human immunodeficiency virus type 1 (HIV-1) is significantly more complex than the genome of simple retroviruses such as ALV. In particular, while ALV encodes three gene products that are translated from only two mRNA species, HIV-1 encodes in excess of twenty mRNAs that permit the efficient translation of nine viral open reading frames (Figure 1). Despite this increased complexity, the basic mechanism remains the same, i.e., alternative splicing in HIV-1 is again regulated by the interplay of cellular transcription factors with suboptimal viral splice sites.21
The mRNAs encoded by HIV-1 can be divided into three classes: (1) the unspliced, ~9 kb transcript; (2) a set of five singly spliced transcripts of ~4 kb, derived by splicing from the major “D1” splice donor to one of the five splice acceptor sites (A1 to A5) located toward the center of the genome (Figure 1); and (3) a complex class of sixteen multiply spliced mRNAs, of ~2 kb in size, derived by exhaustive splicing of the 4 kb class. The 2 kb class encodes the viral regulatory proteins Tat, Rev, and Nef. The 4 kb class encodes the auxiliary proteins Vif, Vpr, and Vpu as well as the viral envelope protein, while the unspliced, genomic length mRNA not only encodes the Gag and Pol proteins but is also packaged into progeny virion particles.
An important distinction among these various viral mRNAs relates to whether they retain functional introns in their primary sequence. In particular, all members of the 2 kb class of viral mRNAs are fully spliced and retain no functional splice donor sequences and, hence, no definable introns. In contrast, both the unspliced 9 kb transcript and the five singly spliced viral RNAs, retain both functional splice donor sites and one or more identifiable introns. It is, of course, the complete removal of these introns that generates the fully spliced ~2 kb class of transcripts.
The reason that this difference is important is schematically illustrated in Figure 2, which shows that capped, polyadenylated mRNAs are normally efficiently and actively transported from the nucleus to the cytoplasm via channels in the nuclear membrane that are termed nuclear pores. However, if an RNA contains an identifiable intron, this induces an interaction with a subset of cellular splicing factors that have been termed commitment factors.15 Although these factors have not been fully defined, they appear to include the U1 small nuclear ribonucleoprotein (snRNP) particle as well as members of the serine-arginine rich (SR) class of splicing factors. For most intron-containing RNAs, this commitment event simply represents the first step in the normal splicing pathway, leading inevitably to the appropriate assembly of further snRNPs and splicing factors on the RNA, to the RNA processing event itself and, once splicing is complete, to nuclear export (Figure 2). Alternatively, if the splice sites are recognized by the commitment machinery but are then found to be nonfunctional, the defective RNA can be degraded within the cell nucleus. Most importantly, however, recognition by commitment factors effectively blocks the nuclear export of the target RNA until and unless the intron(s) present in the RNA are fully removed.3 The purpose of the cellular commitment machinery is, therefore, two-fold. First, these factors function in the identification and definition of introns and, thus, are normally critical for appropriate splicing to occur. Second, these factors effectively retain incompletely spliced cellular RNAs in the nucleus and, thus, prevent pre-mRNAs from encountering the cytoplasmic translational machinery of the cell. Clearly, translation of pre-mRNAs that retain introns within the intended protein coding sequence would generate defective proteins whose expression could be highly deleterious to the cell.

FIGURE 2 While fully spliced RNAs are readily exported from the nucleus, RNAs containing introns are retained in the nucleus by commitment factors, such as the U1 snRNP, until fully spliced or degraded. Rev induces the efficient nuclear export of RRE-containing target RNAs and thereby either prevents or reverses this nuclear retention activity.
THE ROLE OF REV IN THE HIV-1 REPLICATION CYCLE
The existence of the splicing commitment pathway presents retroviruses in general, and HIV-1 in particular, with a serious conundrum. On the one hand, HIV-1 encodes a range of singly and multiply spliced mRNAs whose expression is critical to the viral life cycle. Therefore, the HIV-1 genome must contain splice sites that are recognizable by cellular splicing commitment factors. On the other hand, HIV-1 replication also requires the cytoplasmic expression of unspliced (9 kb) and incompletely spliced (4 kb) transcripts that retain these same splice sites. Therefore, the critical problem is: how do you express RNA transcripts that encode functional splice sites and yet avoid the nuclear retention of unspliced forms of these same transcripts by cellular commitment factors? As indicated in Figure 2, this problem is solved, in the case of HIV-1, by the Rev regulatory protein. As noted above, Rev is one of three viral proteins, along with Tat and Nef, that are encoded by the fully spliced ~2 kb class of viral mRNAs. Because these mRNAs are fully spliced, their nucleocytoplasmic transport and translation is constitutive in expressing cells. Once a significant level of Rev is expressed (this is dependent on the function of the Tat transcriptional transactivator encoded by HIV-1), it acts in the nucleus to induce the nucleocytoplasmic transport of incompletely spliced HIV-1 RNAs that contain the cis-acting Rev response element (RRE) RNA target for Rev.4,18 The RRE is a highly ordered RNA stem-loop structure that is encoded within the body of the HIV-1 env gene (Figures 1 and 3). As such, it is present in all the incompletely spliced viral RNA transcripts whose expression is dependent on Rev.
The phenotype of the HIV-1 Rev protein is clearly revealed in Figure 4, which shows a northern analysis of cytoplasmic poly(A)+ RNA. The cytoplasmic RNA probed in lane 2, derived from cells expressing a wild-type HIV-1 provirus, contains readily detectable levels of all three classes of viral mRNA. In contrast, the cytoplasmic RNA probed in lane 3, derived from a cell expressing an HIV-1 provirus bearing a defective rev gene, contains high levels of the fully spliced 2 kb class of viral transcripts but lacks any evidence for expression of the incompletely spliced 9 kb and 4 kb viral mRNA species. Importantly, published analyses of nuclear viral mRNA expression patterns4,18 clearly demonstrate substantial levels of expression of these 9 kb and 4 kb viral RNAs in the nuclei of cells both in the presence and absence of Rev. There is, therefore, no evidence based on these data to suggest that Rev interferes with splicing directly. Instead, these observations clearly suggest that Rev functions as an RNA sequence-specific nuclear export factor.
More recently, an elegant analysis of Rev function using microinjected Xenopus oocytes has unequivocally demonstrated that Rev can directly induce the nuclear export of target RNAs from the nucleus.6 Of interest, Rev was able to induce the efficient nuclear export of not only RNAs retained by splicing commitment factors, but also RNAs that are inefficiently exported for other reasons, such as the absence of an appropriate 5′ cap structure. Overall, these data have clearly fully validated the earlier proposal that Rev is a sequence-specific nuclear export factor.
Before moving on to a discussion of the mechanism of action of Rev, I would briefly like to discuss the question of how simple retroviruses, such as ALV, promote the nuclear export of their genome length RNA transcript. In principle, in this simpler system one could envision several possible mechanisms. For example, the single-splice donor in ALV could form part of an extended RNA secondary that could randomly fold into two distinct conformations. In one case, the splice donor could be sequestered in an RNA stem, a location known to block recognition by splicing factors. In the second conformation, it could be fully exposed and, hence, efficiently recognized by splicing commitment factors. Although this model remains possible for some simple retroviruses, it is clearly not valid for avian retroviruses such as ALV or for type D retroviruses such as Mason-Pfizer monkey virus (MPMV). Instead, these viruses contain a cis-acting structured RNA target sequence, termed the constitutive transport element (CTE), that functions like the HIV-1 RRE to promote cytoplasmic expr...
Table of contents
- Cover
- Half Title
- Title Page
- Copyright Page
- Table of Contents
- Preface
- Introduction
- Regulation of the Papillomavirus E6 and E7 Oncoproteins by the Viral E1 and E2 Proteins
- The Control of Human Papillomavirus Transcription
- Transcriptional Regulation of HIV
- Role and Mechanism of Action of the HIV-1 Rev Regulatory Protein
- Posttranscriptional Control: A General and Important Regulatory Feature of HIV-1 and Other Retroviruses
- Posttranscriptional Regulation of Papillomavirus Gene Expression
- Questions About RNA Structures in HIV and HPV
- Sequence Redundancy in Biopolymers: A Study on RNA and Protein Structures
- Plurality in HIV Genetics
- Combinatorial Approaches to Inhibiting the HIV Rev-RRE Interaction
- States, Trajectories, and Attractors: A Genetic Networks Perspective of Viral Pathogenesis
- Some Reflections on Complexities in Understanding Virus Biology
- Index