
This is a test
- 352 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Transcription Factors
Book details
Book preview
Table of contents
Citations
About This Book
Transcription factors are important in regulating gene expression, and their analysis is of paramount interest to molecular biologists studying this area. This book looks at the basic machinery of the cell involved in transcription in eukaryotes and factors that control transcription in eukaryotic cells. It examines the regulatory systems that modulate gene expression in all cells, a s well as the more specialized systems that regulate localized gene expression throughout the mammalian organism.
Transcription Factors updates classical knowledge with recent advances to provide a full and comprehensive coverage of the field for postgraduates and researchers in molecular biology involved in the study of gene regulation.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Transcription Factors by Joseph Locker in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Genetics & Genomics. We have over one million books available in our catalogue for you to explore.
Information
1
RNA polymerase II transcription machinery
1.
Introduction: promoter function and strategies in gene control
The set of instructions or genes that define an organism constitutes its genome. The genomes of free-living, single-celled bacteria and eukaryotes contain 4000–6000 genes (Goffeau et al., 1996). In higher eukaryotes, including man, that number can rise to about 70 000. Each gene encodes the information to synthesize a particular protein (via a messenger RNA intermediate) or a particular functional RNA such as transfer RNAs or ribosomal RNAs. Not all genes are turned ‘on’ to the same level or at the same time. While many genes are required to make the common basic constituents of a cell, and thus are constitutively ‘on’, others are induced only when needed to make specific cell types or to respond to a changing environment. Selective gene expression is the end result of a variety of signal transduction pathways that originate from signaling molecules. Both constitutive and inducible gene expression are controlled by the transcriptional regulators, activators and repressors described in the ensuing 11 chapters. DNA recognition occurs through precise molecular complementarity between amino acids in the regulator protein and nucleotide bases in the DNA. A promoter-bound activator then facilitates the recruitment of the transcription machinery to initiate the decoding of the gene. Binding sites for transcriptional regulators, in general, are not confined to precise locations near the transcriptional start site, but may be located as far as 10000 base-pairs upstream or downstream of the transcription start site. However, due to the compaction of DNA in chromatin, these sites may be physically quite close to the transcription start site.
The core of the promoter is composed of one or more DNA elements that are recognized by components of the general transcription machinery, and thus is the assembly site of the transcription machinery. In some cases, multiple core promoters at various locations along a single gene may be employed to provide flexibility in transcribing parts of the gene and generating functionally related but distinct protein products. This chapter will focus on the assembly of the transcription machinery at core promoters. It is important to keep in mind that in vivo transcriptional regulators orchestrate this assembly process.
2.
Architecture of the core promoter
A critical feature of many but not all core promoters is the TATA box, which is recognized by the TATA-binding protein (TBP) subunit of transcription factor, TFIID (Hernandez, 1993). A typical strong TATA box sequence is TATAAAA. RNA transcripts that require a particular TATA box begin 25–30 nucleotides (~60 in yeast) downstream of this element. Genes that are highly transcribed typically possess strong TATA boxes. A weak TATA box might have one or more nucleotide substitutions in the DNA element. A large subset of promoters are ‘TATA-less’ in that they lack any TATA-like consensus sequence in their–30 region, and are generally unaffected by mutations in this region. TATA-less promoters are often, but not always, associated with genes that are transcribed constitutively at low levels.
TATA-less promoters rely largely on an ‘initiator’ element. The initiator was originally characterized as having a pyrimidine (Py) rich consensus: PyPyANA/TPyPy, with the ‘A’ nucleotide being the transcriptional start site (Smale and Baltimore, 1989). This element may be recognized in part by RNA polymerase II (pol II) (Carcamo et al., 1991). More complex initiators have been described, and include additional downstream recognition sequences for gene-specific regulators such as YY1 (yin yang I) (Usheva and Shenk, 1994), TFII–I (Roy et al., 1993), E2F (Means et al., 1992) and upstream stimulatory factor (USF) (Du et al., 1993), as well as components of the core transcription machinery such as the TBP-associated factor (TAF) TAFII150 subunit of TFIID (Verrijzer et al., 1994). Both the TATA box and the initiator may work independently or cooperatively (Emami et al., 1997). Many TATAless promoters also contain a conserved downstream promotor element (DPE) promoter element located approximately 30 nucleotides downstream of the transcriptional start site (consensus: (AG)G(AT)CGTG), which is recognized by the histonelike TAFII60 and TAFII40 subunits of TFIID (Burke and Kadonaga, 1997; 1996).
In vitro, core promoters can drive the efficient expression of an attached gene. However, in vivo, core promoters alone are transcriptionally silent (and do not naturally exist). The difference can be attributed to repression of both the core promoter and components of the transcription machinery in vivo, which are lost upon biochemical fractionation in vitro. In addition to the core promoter, gene expression in vivo is critically dependent upon gene-specific activator sequences or enhancers (Chapter 2). Activators that bind to these elements facilitate the removal of inhibitors as well as directly assisting in the loading of the transcription machinery at the core promoter.
3.
Rate-limiting steps in gene expression
The assembly of a pol II transcription complex can be recapitulated in vitro at a core promoter lacking activators. Such biochemical fractionation and reconstitution of promoter-specific transcription has been instrumental in identifying the general transcription factors, and has provided insight into the assembly pathway and the interacting partners. While biochemical experiments have defined potential mech anistic steps in transcription complex assembly and their regulation, ascertaining whether such steps are rate-limiting for gene expression in vivo is more difficult.
Within the natural state of a cell, some genes are ‘off’, others are transcribed at low levels, while others are fully induced. Any particular gene can move between these states in response to cellular and environmental signals. Consequently, the step that is rate-limiting for the expression of a gene will vary depending upon the incoming signals. In general, genes that are ‘off’ do not have transcription complexes assembled at their promoters, and the rate-limiting step may be the removal of bound repressors by transcriptional activators. Such repressors include histone proteins that wrap DNA into nucleosomes. Some activators function in part by recruiting nucleosome remodeling complexes, which by some unknown mechanism, increases the accessibility of the nucleosomal DNA to the general transcription machinery (Grant et al., 1997).
While accessibility of core promoter DNA is essential for transcription complex assembly, it does not appear to be sufficient. Thus, in general, a transcription complex will not assemble at a core promoter in vivo, even under conditions where the promoter might be accessible. Despite the ability of TBP to recognize the TATA box, it appears to be unable to do so in vivo unless directed by a promoterbound transcriptional activator (Jackson-Fisher et al., 1999; Kuras and Struhl, 1999; Li et al., 1999; Selleck and Majors, 1987). A number of genetic screens have identified mutant proteins that allow high levels of activator-independent transcription, suggesting that TBP and other components of the general transcription machinery are intrinsically repressed (Blair and Cullen, 1997; Prelich and Winston, 1993). Thus a second potential function of activators is to alleviate repressors associated with the general transcription machinery. A third potential function of transcriptional activators is to direct the general transcription machinery to the proper promoter.
4.
Assembly and regulation of the TATA-binding protein and associated factors
TBP is one of the first components of the general transcription machinery to be recruited to the promoter (Figure 1). It is a highly conserved protein found in two of the three domains of life: archaea and eukarya. TBP is unique in that it is required for the expression of all genes, even those transcribed by pol I and pol III. Interestingly, not all genes contain a TATA box, but evidence suggests that TBP nevertheless binds promoter DNA in approximately the same location where the TATA box normally resides (Pugh and Tjian, 1991; Zenzie-Gregory et al., 1993). Apparently, other sequence-specific DNA-binding proteins that directly or indirectly interact with TBP properly position TBP at such TATA-less promoters.
While it is unclear what essential role TBP plays at a promoter, two interesting properties stand out. First, TBP bends DNA by nearly 90°, so it might function to bring transcription factors located distally on either side of TBP together (Kim et al., 1993a; 1993b). Second, TBP binds to a number of the components of the transcription machinery, so it might orchestrate the proper assembly of the transcription machinery.
TBP is a component of a number of functionally distinct multi-subunit complexes that are illustrated in Figure 2 (Hernandez, 1993). Some of these complexes appear to function only with certain RNA polymerases: SL1, TFIID, and TFIIIB are specific for pols I, II, and III, respectively. SNAP appears to function with both pol II and III, but only at a subset of genes, which includes the snRNA genes. Another TBP-containing complex, SAGA (Spt-Ada-Gcn5-acetyltransferase), has important roles in chromatin remodeling. Other TBP complexes, containing factors such as NC2 and Mot1, appear to represent a repressed state of TBP
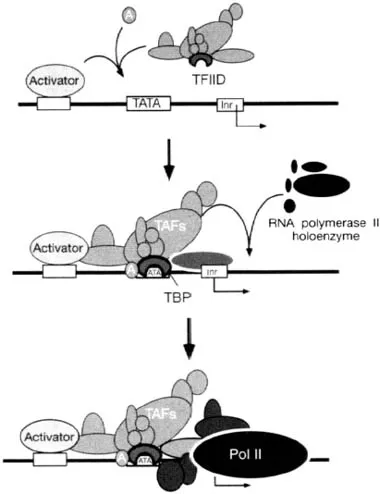
Figure 1. A two-stage model for the assembly of an RNA polymerase II (pol II) transcription complex. A transcriptional activator binds to its cognate site on a promoter and recruits transcription factors TFIID and TFIIA approximately 30 nucleotides upstream from the transcriptional start site. The combined action of this complex recruits the RNA polymerase holoenzyme. Reprinted from Gene 255, Pugh, Control of gene expression through regulation of the TATA-binding protein, 1–14, 2000, with permission from Elsevier Science.
Perhaps the best characterized of the TBP complexes is TFIID, which is recruited to pol II transcribed promoters by transcriptional activators (Figure 1). Apart from TBP, TFIID is composed of about a dozen distinct subunits that are referred to as TAFIIs. TAFIIs fall into a variety of categories. First, some TAFIIs form structures akin to histones, and like histones may assemble into a nucleo-some-like structure containing two copies each of the four different histone-like TAFIIs (Xie et al., 1996). Second, a number of the TAFIIs, including the histone-like TAFIIs, appear to reside in the SAGA complex, and thus both SAGA and TFIID are likely to share some common functions (Grant et al., 1998; Imhof et al., 1997; Martinez et al., 1998). Third, some TFIID TAFIIs appear to have either structural or functional homologs in SAGA. For example, the TAFII28/TAFII18 subunits have a s...
Table of contents
- Front Cover
- Half Title
- Title Page
- Copyright
- Contents
- Contributors
- Abbreviations
- Preface
- 1 RNA polymerase II transcription machinery
- 2 Regulatory transcription factors and cis-regulatory regions
- 3 Chromatin structure and regulation of transcription
- 4 DNA binding by transcription factors
- 5 Regulation of transcription during the cell cycle
- 6 Regulation of transcription by extracellular signals
- 7 Transcriptional regulation of early lymphocyte development
- 8 Transcriptional integration of hormone and metabolic signals by nuclear receptors
- 9 Developmental control by Hox transcriptional regulators and their cofactors
- 10 Tissue-specific regulation by transcription factors
- 11 Neural transcription factors
- 12 Abnormal transcription factors produced by chromosome translocations in human cancer
- Index