
- 299 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
DNA Microarrays
About this book
DNA Microarrays introduces all up-to-date microarray platforms and their various applications. It is written for scientists who are entering the field of DNA microarrays as well as those already familiar with the technology, but interested in new applications and methods.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
1 Introduction: DNA microarrays – ten years old, but no old hat
Falk Hertwig and Ulrike A Nuber
In October this year, we celebrate the 10th anniversary of DNA microarrays, as this technology was first mentioned in an article by Schena, M., Shalon, D., Davis, R.W., and Brown P.O. published in the ‘Genome Issue’ of Science in 1995 (1). Predecessors of this technology were dot blots, slot blots, and macroarrays with membranes used as platform. Microarrays have been fascinating the scientific community for the last decade, but still have not reached the limits of their potential and are continuing to invade new fields of biology and medicine.
In the course of deciphering the genomes of many organisms, the need for functional studies of thousands of genes arose, and one step towards this goal has been the identification of expression patterns of genes under normal and pathological conditions. Coincidentally, the ‘Genome Issue’ of Science in 1995 contained two articles that describe mRNA profiling at a large scale: the DNA microarray paper by Schena et al. (1), and the paper by Velculescu et al. (2) on SAGE (serial analysis of gene expression). However, it seems that microarrays ‘won the race’ in the field of gene expression profiling – which is mainly due to the high throughput (number of investigated samples per time), and rapid readout of the results.
Schena et al. (1) used their first microarray to monitor gene expression – which is undeniably still the most prominent application – but the possibility to use this technique for other purposes is only limited by the scientist’s creativity and budget.
This book focuses on microarrays that consist of immobilized DNA molecules, but similar miniaturized hybridization formats have been developed, such as lipid microarrays (3) and protein microarrays (for review see 4). The latter can, for example, be used to monitor the interactions of immobilized proteins with proteins, nucleic acids and small molecules, and offers applications in medical diagnostics (e.g. the screening of patients’ sera for specific antigenic properties) (5), and in basic research (e.g. the identification of ligands using protein receptor arrays) (6).
There is probably no field in life sciences (basic or applied) which has not been impinged upon by DNA microarrays. Some examples are their application in cancer research (Chapter 4), pharmacogenomics (Chapter 2), and stem cell research (Chapter 5). In theory, DNA microarrays of every organism can be generated and used, dependent soley on the availability of DNA sequence information. In this book, the use of yeast, plant, mouse, rat and human arrays is described (see Figure 1.1 ).
Apart from monitoring gene expression, DNA microarrays are used to detect single nucleotide changes (Chapters 6, 7, and 8), unbalanced chromosome aberrations by array CGH (Chapters 11 and 12), or balanced chromosome aberrations by array painting (Chapter 12). The ChIP-on-chip technology was established only a few years ago as a new microarray tool enabling the search for transcription factor binding sites at a large scale (Chapters 13 and 14).
Strangely enough, there are still some scientists who are reluctant to use DNA microarrays. ‘With microarrays, you see too much!’ ‘But there are so many differentially expressed genes…’ – If this sounds like you, don’t be shy, read on. Chapters in this book will guide you through the processing of complex data and introduce you to different approaches for handling your results.
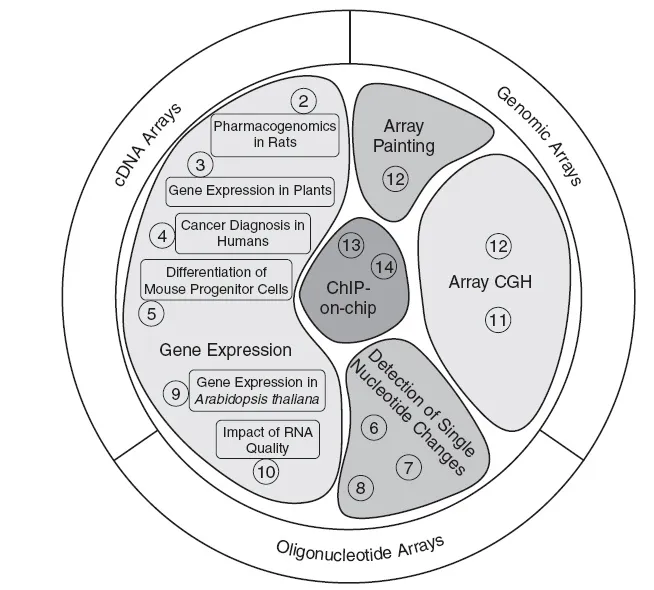
Figure 1.1.
Chapter overview showing different microarray platforms and applications described in this book. Numbers indicate book chapters.
In contrast, other scientists, seduced by the temptation to produce vast amounts of data, may face some of the pitfalls associated with DNA microarray experiments. If you belong to this category, chapters in this book will show you some strategies on how to set up a good microarray experiment, resulting in relevant data, and how to avoid producing data which cannot be processed by the best bioinformatician…
Finally, to the rest of the interested readers, who may have already gained some experience with DNA microarrays, this book should provide new aspects (for example the impact of RNA quality on Affymetrix GeneChip analyses, Chapter 10) and detailed protocols covering all steps of a microarray experiment from the production of the array to data analysis and storage.
1.1 How to perform a microarray experiment
While musing at the thought of using microarrays, many questions pop up in one’s mind: What type of arrays do I want to use? How do I design my experiments? Which labeling method do I want to use? How do I analyze my microarray data?
To obtain relevant results and perform a microarray experiment that best suits your needs, you should carefully set up a plan before starting at the bench. You will find help and inspiration in the various chapters. Regarding the design of DNA microarray experiments, we would like to refer to a review by Yang and Speed (7).
DNA microarrays can be classified according to the type of probes on the array (cDNA, oligonucleotides, genomic fragments), their generation and immobilization. In many cases presynthesized molecules (PCR products, oligonucleotides, isolated DNA) are deposited on the array either by contact printing (using metal pins that carry small volumes of probe solution due to capillary action) or by non-contact printing, when probe solution is dispensed by ink-jet printing. In addition, several companies generate high-density microarrays by synthesizing oligonucleotides in situ (for review see 8). The synthesis is either based on specific base deprotection by light (coordinated by photomasks or digital micromirror devices) or on chemical deprotection and the use of ink-jet technology. An alternative to these commercial systems is the open-source platform POSaM (piezoelectric oligonucleotide synthesizer and microarrayer), described by Lausted et al. (9). These authors present the low-cost production of in situ synthesized oligonucleotide arrays (containing 9800 features) in their lab. An overview of commercially available oligonucleotide arrays is given in Table 1.1.
cDNA arrays can also be purchased (see Table 1.2), or produced as described in various chapters of this book (Chapters 2, 3, 4, and 5). In addition to protocols for the generation of genomic DNA arrays (see Chapters 11 and 12), you can find commercial suppliers in Table 1.2 .
After the hybridization of labeled target molecules to DNA arrays, fluorescent signals are detected by laser scanners or CCD cameras. Chapters 15 and 16 describe image acquisition and image data conversion.
Finally, microarray data produced in large scale need to be stored and processed to obtain relevant and meaningful results. At this point the field of bioinformatics found a vast playground and the increased use of microarrays triggered the co-evolution of various bioinformatics methods (Figure 1.2). In general, all data need to be normalized. Not to get you lost at this early step, several normalization methods are presented in Chapter 17. The special case of normalizing array CGH data is described in Chapter 21.
Table 1.1. Commercially available oligonucleotide microarrays
Table 1.2. Companies producing cDNA and genomic microarrays
After normalization, further data processing depends on the questions you intend to address. If you want to compare gene expression under different biological conditions (e.g. normal vs. tumor tissue, wild type vs. knockout/transgene cells, control vs. drug-treated cells etc.) the first and foremost question concerns differential gene expression. Chapter 18 takes you on a safe trip to a list of differentially expressed genes.
DNA microarray time course experiments are highly suitable to monitor the expression of a very large number of genes during a biological process over a defined period of time (one example is given in Chapter 5). Chapter 20 describes the statistical analysis of time-course data.
Chapter 19 deals with clustering and classification. Clustering is, for example, used to find a group of genes, which have similar expression patterns or a group of samples (e.g. tissue samples from patients), which show likewise expression of a set of genes. Classification methods can determine whether a gene expression profile of a tissue sample belongs to a certain class, and are applied to predict disease courses.
Finally, special applications require special bioinformatic analyses. Therefore, the processing of data generated by single nucleotide polymorphisms (SNP) detection and ChIP-on-chip experiments is addressed separately in Chapters 8 and 13, respectively.
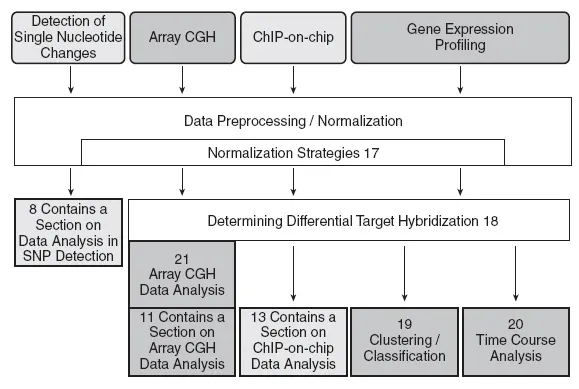
Figure 1.2.
Flowchart of bioinformatics methods that are used at different steps of microarray data analysis. Numbers indicate book chapters.
Ten years ago, DNA microarrays became fashionable because they enable high-throughput gene expression analyses. Over the years, they gained importance with their expanding use in medical diagnostics and research and even now scientists are continuing to advance this technology and its applications. The examples of ChIP-on-chip and array painting show that the combination of two techniques can lead to a new technology – so it will be exciting to see what’s yet to come….
References
1. Schena M, Shalon D, Davis RW and Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470.
2. Velculescu VE, Zhang L, Vogelstein B and Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484–487.
3. Lahiri J, Jonas SJ, Frutos AG, Kalal P and Fang Y (2001) Lipid microarrays. Biomed Microdevices 3: 157–164.
4. Labaer J and Ramachandran N (2005) Protein microarrays as tools for functional proteomics. Curr Opin Chem Biol 9: 14–19.
5. Lueking A, Possling A, Huber O, Beveridge A, Horn M, Eickhoff H, Schuchardt J, Lehrach H and Cahill DJ (2003) A nonredundant human protein chip for anti-body screening and serum profiling. Mol Cell Proteom 2: 1342–1...
Table of contents
- Cover Page
- Title Page
- Copyright Page
- Abbreviations
- 1 Introduction: DNA microarrays – ten years old, but no old hat
- 2 cDNA microarray analysis and its role in toxicology – a case study
- 3 Gene expression profiling in plants using cDNA microarrays
- 4 Identification of gene expression patterns for a molecular diagnosis of kidney tumors
- 5 Gene expression analysis of differentiating neural progenitor cells – a time course study
- 6 A microarray-based screening method for known and novel SNPs
- 7 From gene chips to disease chips – new approach in molecular diagnosis of eye diseases
- 8 Multiplexed SNP genotyping using allelespecific primer extension on microarrays
- 9 Profiling the Arabidopsis transcriptome
- 10 Affymetrix GeneChip analyses – the impact of RNA quality
- 11 Molecular karyotyping by means of array CGH: linking gene dosage alterations to disease phenotypes
- 12 DNA microarrays: analysis of chromosomes and their aberrations
- 13 Mapping transcription factor binding sites using ChIP-chip – general considerations
- 14 ChIP-on-chip: searching for novel transcription factor targets
- 15 Turning photons into results: principles of fluorescent microarray scanning
- 16 Microarray detection with laser scanning device
- 17 Normalization strategies for microarray data analysis
- 18 Microarray data analysis: Differential gene expression
- 19 Clustering and classification methods for gene expression data analysis
- 20 Statistical analysis of microarray time course data
- 21 Array CGH data analysis
- 22 MIAME
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access DNA Microarrays by Ulrike Nuber in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Biology. We have over one million books available in our catalogue for you to explore.