eBook - ePub
Cyclic Nucleotide Phosphodiesterases in Health and Disease
This is a test
- 728 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Cyclic Nucleotide Phosphodiesterases in Health and Disease
Book details
Book preview
Table of contents
Citations
About This Book
Since the last major compendium dedicated to cyclic nucleotide phosphodiesterases (PDEs) was published over 15 years ago, an enormous amount of progress has occurred in the field. There is great need for a centralized source for key information in this burgeoning and therapeutically important area of medical research.
Cyclic Nucleotide Phosph
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Cyclic Nucleotide Phosphodiesterases in Health and Disease by Joseph A. Beavo,Sharron H. Francis,Miles D. Houslay in PDF and/or ePUB format, as well as other popular books in Medizin & Biochemie in der Medizin. We have over one million books available in our catalogue for you to explore.
Information
Section A
Specific Phosphodiesterase Families—Regulation, Molecular and Biochemical Characteristics
3 | Calmodulin-Stimulated Cyclic Nucleotide PhosphodiesterasesAndrew T. Bender |
CONTENTS
3.1 Introduction
3.2 Biochemical Characteristics
3.2.1 Primary Structure and Activation by Ca2+/CaM
3.2.2 Kinetic Properties
3.2.3 Regulation by Phosphorylation
3.2.4 Inhibitors
3.3 PDE1 Genetics
3.3.1 PDE1A
3.3.2 PDE1B
3.3.3 PDE1C
3.4 Tissue-Specific Expression and Functional Roles of PDE1
3.4.1 Cardiovascular System
3.4.2 Nervous System
3.4.3 Immune System
3.4.4 Testis and Sperm
3.5 Conclusions
Acknowledgments
References
3.1 INTRODUCTION
The PDE1 family of enzymes has a relatively long history and was one of the first PDE families to be discovered.1,2 It is one of the most studied and well characterized of the 11 PDE families. PDE1 enzymes were initially isolated from animal tissues, particularly bovine, and characterized by their unique biochemical properties. Calmodulin-stimulated PDE activity was originally identified in bovine1,3 and rat brain.2 Subsequent work found that PDE1 enzymes hydrolyze both cAMP and cGMP and distinguished them from other PDEs as having the unique property of being activated by the binding of a complex of calcium and calmodulin (CaM). To date, three PDE1 isoforms PDE1A, PDE1B, and PDE1C have been discovered and are the products of separate genes. The advent of the molecular biology revolution facilitated the cloning and study of the different PDE1 isoforms. Molecular biology advances have also allowed the discovery of the many variants of the three PDE1 genes and aided the study of their expression and localization.
An irksome difficulty of the PDE1 research field has been cryptic nomenclature of the enzymes. Initially, PDEs were classified based on their regulatory properties and PDE1 was simply referred to as CaM-stimulated PDE as researchers were unaware of the multitude of families that were yet to be discovered. As new isoforms were discovered, they were named based on their molecular mass and tissue of discovery, i.e., PDE1B was the “63 kDa bovine brain CaM-stimulated PDE”. The adoption of the current PDE nomenclature system in 19954 has alleviated much of the confusion and instituted a nomenclature system based on PDE primary structure. The current nomenclature scheme uses the first two letters to represent the species, the next three letters plus an Arabic numeral to designate the cyclic nucleotide phosphodiesterase gene family, the next letter to represent the individual gene product within the family, and the final Arabic numeral to represent the variant. Thus, the “63 kDa bovine brain CaM-stimulated PDE” is now identified as BtPDE1B1. In this chapter, separate gene products will be referred to as isoforms and individual mRNA or protein products of the different genes arising from alternative splicing or alternative transcriptional starts will be called variants.
As with many of the PDE families, little is known about the physiological function of the PDE1 enzymes. Study of the function of PDE1 has been hindered by the lack of a readily available PDE1-selective inhibitor that will rapidly enter cells. However, a great deal of work has described the expression pattern of PDE1s in different tissues and cell types and how the expression can change in the course of physiological and pathological processes. These data together with some recent genetic approaches for studying PDE1 have yielded clues about PDE1 function. This chapter will provide only a brief review of the biochemical characteristics of the PDE1s, as their characteristics have been well reviewed previously.5–7 Instead, the main emphasis will be genetic regulation and tissue-specific expression of PDE1 as these areas have been the focus of recent research. Additionally, what has been discovered about the functional roles of PDE1 will be discussed.
3.2 BIOCHEMICAL CHARACTERISTICS
3.2.1 PRIMARY STRUCTURE AND ACTIVATION BY CA2+/CAM
As is common with most PDE families, the PDE1 enzymes have a primary structure consisting of a somewhat conserved C-terminal catalytic domain and an N-terminal regulatory domain. The unique distinguishing feature of the PDE1 family is the existence of two regulatory CaM-binding sites at the N-termini of the enzymes. The enzymes are known to exist as dimers, 8, 9, 10 although the nature or functional significance of dimerization has not been elucidated. The molecular weight of the enzymes ranges from 58 to 86 kDa per monomer.
The binding of an activated Ca2+/CaM complex serves to stimulate PDE1 enzymatic activity. The activation by CaM is an effect on Vmax and not on Km.11 The domain organization of the enzymes includes a C-terminal catalytic domain, two N-terminal CaM-binding domains, and an inhibitory domain situated between the two CaM-binding sites12 (see Figure 3.1A). The inhibitory domain is conserved in all the PDE1 enzymes13 and may interact with a site near the catalytic domain that maintains the enzyme in an inactive conformation. It was demonstrated with PDE1A that upon binding of Ca2+/CaM a conformational change occurs where the inhibition is released and full enzyme activity is allowed (Figure 3.1B).12 CaM-mediated displacement of an autoinhibitory domain has been shown to be an activation mechanism for other proteins such as CaM kinase II, myosin light chain kinase, CaM kinase kinase, and calcineurin.14 However, exactly how the inhibitory domain interacts with the rest of the protein to prevent PDE1 enzyme activity has not been elucidated.
The different PDE1 isoforms vary in sensitivity to activation by Ca2+/CaM (see Figure 3.2). In the presence of excessive CaM, the EC50 for activation by Ca2+ varies from 0.27 μM (PDE1A1) to 3.02 μM (PDE1C1).15 However, the affinity is not uniquely high or low for all the variants of each isoform. Variation in amino acid sequence at the N-terminus of some isoforms confers sequence changes to the first CaM-binding site that can alter PDE1 affinity for CaM. For instance, the PDE1A1 variant has an N-terminus that is distinct from that of PDE1A2 and consequently has a nearly 10-fold lower EC50 for the activation by CaM. These properties produce an array of PDE1 enzymes that would respond to different degrees to changes in Ca2+ levels in the cell. Thus, different cell types may express individual PDE1 enzymes to regulate specific calcium-mediated processes. Additionally, phosphorylation is a posttranslational mechanism that can reversibly decrease the affinity for Ca2+/CaM (Figure 3.2). The variations in Ca2+/CaM affinity produced by phosphorylation and expression of unique N-terminal sequences are likely to be important mechanisms for imparting a highly specific regulation of PDE1 activity by Ca2+ in vivo and the two concepts will be discussed in more detail in later sections. Exactly how the biochemical properties of PDE1 regulation are related to the regulation of the enzyme by Ca2+ signals in intact cells is beginning to be investigated.16
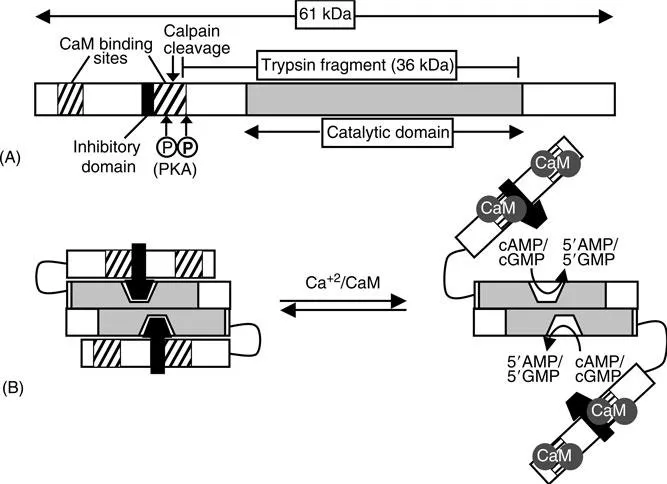
Wide variations in the fold stimulation of PDE1 activity by CaM have been reported. One reason for the variable activation is that proteolysis can cleave off the inhibitory domain thereby irreversibly activating the enzyme. Limited proteolysis of the 59 kDa bovine brain PDE1 in vitro by α-chymotrypsin produced a 45 kDa fragment that could not be activated by the addition of CaM, but had activity equal to native enzyme in the presence of CaM.11 Similarly, cleavage of PDE1A2 with trypsin was used to generate a 36 kDa fragment that was fully active in the absence of CaM.17 By amino acid sequence analysis, the identity of the fragment was shown to contain the catalytic domain (see Figure 3.1A). The proteolytic activation may be phys...
Table of contents
- Cover
- Title Page
- Copyright Page
- Preface
- Editors
- Contributors
- Table of Contents
- Introduction
- Section A Specific Phosphodiesterase Families—Regulation, Molecular and Biochemical Characteristics
- Section B Nonmammalian Phosphodiesterases
- Section C Phosphodiesterases Functional Significance: Gene-Targeted Knockout Strategies
- Section D Compartmentation in Cyclic Nucleotide Signaling
- Section E Phosphodiesterases as Pharmacological Targets in Disease Processes
- Section F Development of Specific Phosphodiesterase Inhibitors as Therapeutic Agents
- Index