
eBook - ePub
Enzymes in the Environment
Activity, Ecology, and Applications
This is a test
- 640 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Enzymes in the Environment
Activity, Ecology, and Applications
Book details
Book preview
Table of contents
Citations
About This Book
Exploring the role of enzymes in biogeochemical processes, this reference explores the function, molecular biology, and biochemistry of microorganisms and their intra-and extra-cellular enzymes in soils and aquatic systems. With contributions from international experts, the book provides detailed discussions on the use of enzymes to assess nutrient turnover, soil health, and environmental stresses. Topics include methods for determining and manipulating the diversity of microbial populations, the effect of biofilms and their microbes ad enzymes on the environment, microbe-plant symbiosis, microbial activities in lake and ocean ecosystems, and terrestrial ecosystem stress, and more.
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access Enzymes in the Environment by Richard G. Burns, Richard P. Dick, Richard G. Burns, Richard P. Dick in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Environmental Science. We have over one million books available in our catalogue for you to explore.
Information
1
Enzyme Activities and Microbiological and Biochemical Processes in Soil
Paolo Nannipieri
Universitá degli Studi di Firenze, Firenze, Italy
Ellen Kandeler
University of Hohenheim, Stuttgart, Germany
Pacifico Ruggiero
Universitá degli Studi di Bari, Bari, Italy
I. INTRODUCTION
It is well known that soil organisms, particularly microbiota, play an essential role in the cycling of elements and stabilization of soil structure (1,2). The mineralization of organic matter is carried out by a large community of microorganisms and involves a wide range of metabolic processes. For this reason, it is important to relate ecosystem structure and function to species and functional diversity (3). However, the relationships between genetic diversity and taxonomic diversity are not well understood and even less is known about the manner in which these two properties affect microbial functional diversity (3– 5). Microbial species diversity is related to richness (i.e., the number of different species), evenness (i.e., the relative contribution that individuals of all species make to the total number of organisms present), and composition (i.e., the type and relative contribution of particular species present) (3). According to the well-known and much used BIOLOG approach, microbial functional diversity is related both to the rates of substrate utilization and to the presence or absence of utilization of specific substrates. A decrease in microbial diversity may reduce microbial functionality of soil if “keystone species,” such as nitrifiers and nitrogen-fixing microorganisms, are negatively affected (6). However, this is not generally the rule because rarely are there only a few species that perform a singular function. Several processes, such as organic carbon mineralization, are carried out by a large number of microbial species and a reduction in any group of species has little effect on overall soil processes since other organisms can fulfill these functions (7– 9). This spare capacity or resilience is a feature of most soil ecosystems.
The molecular techniques used today have identified myriad microbial genes. However, the ecological importance of these genes is largely unknown because it is difficultto quantify their biochemical and microbial expression in situ (10). One significant advance using comparatively recent developments in molecular techniques (based on deoxyribonucleic acid [DNA] extraction, purification, amplification, and analysis) has allowed recording and monitoring the so-called nonculturable microorganisms (11). However, the determination of microbiological activities requires detecting metabolic gene transcripts (messenger ribonucleic acids [mRNAs]) in conjunction with modern sensitive assays of metabolites and mRNA translation products, such as enzymes. These approaches are rapidly improving our understanding of the distribution and extent of microbe-mediated processes in the field. It has been proposed that the monitoring of enzyme activities can be used to determine the effect of genetically modified microorganisms on soil metabolism (12,13).
Enzymes are proteins many of whose activities can be measured in soil. The assays of soil enzymes are generally simple, accurate, sensitive, and relatively rapid. A range of enzyme activities, and a large number of samples, can be analyzed over a period of a few days using small quantities of soil. It is well known that changes in enzyme activities depend not only on variations of gene expression but also on changes of environmental factors affecting the considered activity (14,15). The expression of genes may occur in natural samples, but numerous factors might effectively prevent the actual enzyme process from taking place (Fig. 1). It has been hypothesized that the microbial composition of a soil determines its potential for substrate catalysis since most of the processes occurring in soil are microbe-mediated and are carried out by enzymes (14).
The objective of this chapter is to discuss the potential of soil enzyme assays to determine soil microbial functional or process diversity and, when possible, identify future research needs. The carbon substrate utilization approach is compared with enzyme measurements for monitoring soil microbial functional diversity. Since soil microbial functional diversity encompasses several metabolic activities, this approach requires assays of many hydrolytic and oxidative enzymes. Therefore, the discussion includes the use of composite indices or multivariate statistical analysis for integrating enzyme data sets for comparing soil samples. Methods that distinguish between the contributions of extracellular and intracellular enzyme reactions are discussed.
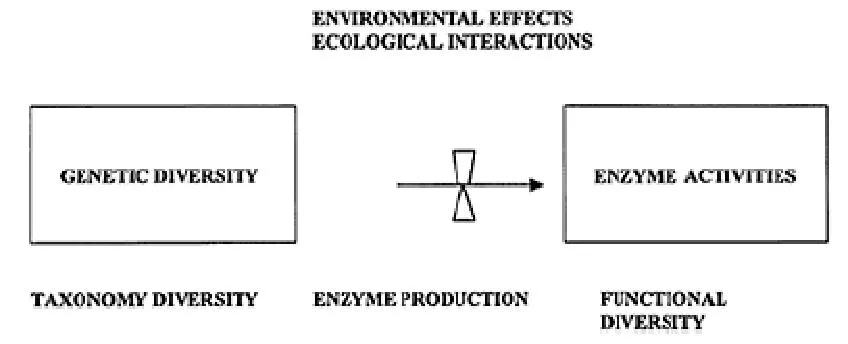
Figure 1 Scheme showing the possible relations among taxonomic diversity, genetic diversity, functional diversity, and enzyme activity. (From Ref. 14.)
II. CARBON SUBSTRATE UTILIZATION PATTERNS AND MICROBIAL FUNCTIONAL DIVERSITY
The BIOLOG system, originally developed to assist with the taxonomic description of bacteria in axenic culture, is based on the ability of bacterial cells to oxidize up to 95 different carbon substrates in microtiter plates (16). The plates are incubated for a suitable period of time (generally 72 h), and the oxidation of the substrate is monitored by measuring the reduction of a tetrazolium dye. The rate and density of the color change depend upon the number and activity of microbial cells in the well of the microtiter plate. The BIOLOG system also has been applied to assess the functional metabolic diversity of microbial communities from different habitats (17) and soil types (18–20), including the rhizosphere (21,22) and grassland soils (23). Microbial communities produced habitatspecific and reproducible patterns of carbon source oxidation, and thus this technique was suggested to be sensitive for detecting temporal and spatial differences among soil microbial communities (24). The carbon substrate utilization profiles have been shown to be sensitive to heavy metal pollution (25), organic pollutants (26), soil types (26,27), crop type, and crop management (28). The last mentioned study showed that, of the expectants tested, 0.01 M NaCl yielded the highest well color development (28).
The BIOLOG approach presents several advantages. First, there is no doubt that the utilization of carbon is a key factor in governing microbial growth in soil, and, for this reason, this technique has been considered ecologically important (Table 1). In addition, the technique is very rapid and simple. Thus it has become a very popular method of assessing soil metabolic diversity and therefore soil functionality. In 1997 an international meeting was devoted almost completely to the subject (29).
However, there are several drawbacks that limit the accuracy of the method in assessing the metabolic diversity of soil microbiota (Table 1). First, the inoculum is a mixture of organisms extracted from soil rather than a single species from axenic culture. Thus, only the culturable minority of microorganisms is capable of oxidizing the organic substrates (30). It is well established that only 1–10% of soil microflora are culturable by a range of conventional techniques and media (31). Furthermore, reproducible results can be obtained only if replicates present a similar inoculation density (24). Because BIOLOG plates were developed for a different type of microbiological analysis, not all of the 95 organic substrates offered are ecologically relevant, and it could be important to choose organic compounds that are appropriate for the microorganisms in their specific habitats. For example, organic carbon compounds commonly present in root exudates were tested to discriminate between the functional activities of microbial communities in rhizosphere soil (32). Insam (33) reduced the number of substrates to 31 of those reported in soil to allow three replicates of each substrate (plus a control) in a 96-well plate. Another problem is that the multivariate statistics used (principal component analyses, discriminant analyses, and detrended correspondence) to analyze BIOLOG data may mask the analytical problems described. Changes in the microbial composition of the inoculum may occur during the incubation (Table 1). Indeed, not all constituents of the inoculum contribute to the color development, and significant changes in the community structure occur over a 72-h period (34). By comparing DNA melting profiles (denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis) at the beginning and at the end of the incubation, it was shown that the structure of the microbial community changed in potato rhizosphere soil. In particular, it was observed that fast-growing bacteria become dominant during the incubation period. However, the changes were not observed in an activated sludge reactor amended with glucose or peptone (34). It was postulated that in this case the dominant bacteria had been selected for rapid growth on readily utilizable carbon sources because the activated sludge had been continuously fed with glucose and peptone. In contrast, the dominant bacterial population of the potato rhizosphere was selected in an environment characterized by a much lower content of organic substrates than in activated sludge. Therefore, the dominant bacterial population (equivalent to K strategists) of the rhizosphere inoculum was displaced by a more competitive microbial population (equivalent to r strategists). Another significant limitation is that the community profiles obtained with the use of the BIOLOG procedure do not include fungi because of their comparatively slow growth rate (19).
Table 1 Advantages and Disadvantages of the BIOLOG Technique
Degens and Harris (35) have attempted to overcome the many problems of the BIOLOG approach by measuring the patterns of in situ catabolic potential of microbial communities. Differences in the individual short-term respiration responses (or substrate induced respiration [SIR]) of soils to the addition to 36 simple substrates were used to assess microbial communities. Despite the fact that this approach seems more accurate than the BIOLOG technique, it has not been used widely since it was first published in 1997.
III. ENZYME ACTIVITIES AND SOIL MICROBIAL FUNCTIONAL DIVERSITY
A. Limitations of a Single Enzyme Activity as an Index of Microbial Activity
Enzyme activities can be measured and used as an index of microbiological functional diversity if they reflect changes in microbial activities. Since microbial functional diversity includes many different metabolic processes, theoretically a large number of different enzymes should be measured. Data should be integrated in an attempt to calculate an index of microbial functional diversity and to compare the microbial functional diversity of different soil samples. Because this task is not possible, a representative set of enzyme activities is needed. One approach might be to measure only the enzyme activities that control the key metabolic pathways. Generally, these are rate-limiting exergonic steps and are the targets of metabolic regulation. In the case of glycolysis (one of the main metabolic pathways present in almost every microbial cell and transforming 1 mole of glucose to 2 moles of pyruvate), phosphofructokinase 1 is one of the 10 enzymes involved and catalyzes the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate. This regulates the rate of glycolysis (36). Therefore, determination of the phosphofructokinase-1 activity should be an indication of the potential rate of glycolysis in soil...
Table of contents
- Cover Page
- Title Page
- Copyright Page
- Preface
- Contributors
- 1: Enzyme Activities and Microbiological and Biochemical Processes in Soil
- 2: Ecology of Microbial Enzymes in Lake Ecosystems
- 3: Ecological Significance of Bacterial Enzymes in the Marine Environment
- 4: Enzymes and Microorganisms in the Rhizosphere
- 5: Enzymes in the Arbuscular Mycorrhizal Symbiosis
- 6: Microbes and Enzymes Associated with Plant Surfaces
- 7: Microbial Enzymes in the Biocontrol of Plant Pathogens and Pests
- 8: Microbiology and Enzymology of Carbon and Nitrogen Cycling
- 9: Enzyme and Microbial Dynamics of Litter Decomposition
- 10: Fungal Communities, Succession, Enzymes, and Decomposition
- 11: Enzyme Adsorption on Soil Mineral Surfaces and Consequences for the Catalytic Activity
- 12: Microbes and Enzymes in Biofilms
- 13: Search for and Discovery of Microbial Enzymes from Thermally Extreme Environments in the Ocean
- 14: Molecular Methods for Assessing and Manipulating the Diversity of Microbial Populations and Processes
- 15: Bioindicators and Sensors of Soil Health and the Application of Geostatistics
- 16: Hydrolytic Enzyme Activities to Assess Soil Degradation and Recovery
- 17: Enzymatic Responses to Pollution in Sediments and Aquatic Systems
- 18: Microbial Dehalogenation Reactions in Microorganisms
- 19: Isolated Enzymes for the Transformation and Detoxification of Organic Pollutants
- 20: Enzyme-Mediated Transformations of Heavy Metals/Metalloids: Applications in Bioremediation
- 21: Enzymes in Soil: Research and Developments in Measuring Activities