
eBook - ePub
PCR Guru
An Ultimate Benchtop Reference for Molecular Biologists
This is a test
- 104 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Book details
Book preview
Table of contents
Citations
About This Book
PCR Guru: An Ultimate Benchtop Reference for Molecular Biologists is provides researchers in molecular biology with a handy reference for approaching and solving challenging problems associated with PCR setup and optimization. As a laboratory guide, it emphasizes the technical aspects of employing PCR as a tool in molecular biology laboratories. The book covers the history of PCR and the basic science underlying it. It then discusses PCR at the bench level, starting with detailed description and tips on primer design, and continuing with the standard protocols used to perform PCR.
- Provides troubleshooting tips for various types of modifications of standard protocols
- Contains unique "Good Practices and Tips" that are indispensable for the beginner and expert alike
- Features "Special Cases" with applications of PCR, optimization, and troubleshooting
- Includes detailed appendices with tables, figures, and key protocols
- Organized as a systematic, concentrated resource to save time when addressing a PCR problem
Frequently asked questions
At the moment all of our mobile-responsive ePub books are available to download via the app. Most of our PDFs are also available to download and we're working on making the final remaining ones downloadable now. Learn more here.
Both plans give you full access to the library and all of Perlego’s features. The only differences are the price and subscription period: With the annual plan you’ll save around 30% compared to 12 months on the monthly plan.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 1000+ topics, we’ve got you covered! Learn more here.
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more here.
Yes, you can access PCR Guru by Ayaz Najafov,Gerta Hoxhaj in PDF and/or ePUB format, as well as other popular books in Biological Sciences & Genetics & Genomics. We have over one million books available in our catalogue for you to explore.
Information
Chapter 1
Introduction
Abstract
Origins of polymerase chain reaction (PCR) and its history are briefly introduced in this chapter. The components of the PCR are introduced and the effect of the component concentrations on the fidelity, specificity, and efficiency of PCR are explained. Various thermostable DNA polymerases that are currently being used in the PCR setups around the world are listed. The power of the cyclic and the exponential nature of PCR, the effect of the reaction reaching the plateau, and how this affects the specificity and yield of the reaction are discussed. Finally the rise in the popularity of this method since it was first introduced by Dr. Kary Mullis in 1983 is illustrated.
Keywords
PCR
history
Kary Mullis
components
fidelity
specificity
efficiency
exponential
polymerases
For knowledge itself is power.
(Francis Bacon)
Every great advance in science has issued from a new audacity of imagination.
(John Dewey)
Real knowledge is to know the extent of one’s ignorance.
(Confucius)
1.1. A bit of history
Polymerase chain reaction (PCR) was invented by Dr. Kary Mullis in 1983. At that time, he was working at Cetus Corporation, one of the first biotechnology companies. For his invention, Dr. Mullis received a $10,000 bonus from Cetus. In 1992, Dr. Mullis sold the patent for PCR and Taq polymerase to Hoffmann La Roche for $300 million. In 1993, he received a Nobel Prize in Chemistry “for his invention of the polymerase chain reaction method” [1]. To read more about the history of PCR see Ref. [2].
1.2. What is PCR?
It is a rapid, relatively inexpensive, simple, sensitive, versatile, and robust in vitro method for specifically amplifying desired sequences of DNA or RNA molecules (Fig. 1.1). A large amount of products (up to milligrams of double-stranded DNA fragments) are generated at the end of each PCR, and a huge variety of downstream experiments and applications have been invented due to this technique’s ability to generate double-stranded DNA at a large scale (Fig. 1.2). The applications of PCR include classification of organisms, genotyping, molecular archaeology, mutagenesis, mutation detection, sequencing, cancer research, detection of pathogens, DNA fingerprinting, drug discovery, genetic matching, genetic engineering, and prenatal diagnosis. The high applicability of PCR comes from its specificity and ability to detect even a single DNA molecule and produce millions of copies of the desired target sequence. As it is very hard, if not impossible, to manipulate DNA molecules at low quantities, by amplifying the DNA quantities to the levels that are very easy to handle, PCR revolutionized how people think about DNA experiments.
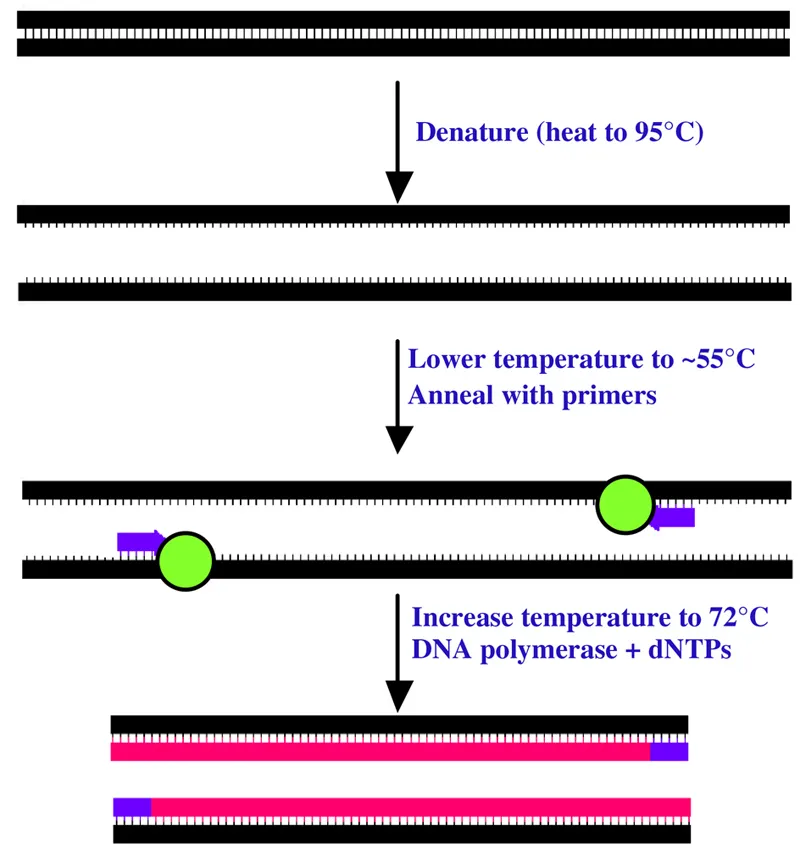
Figure 1.1 Steps during the polymerase chain reaction (PCR).
DNA polymerase (green), primers (purple), and dNTPs (pink) are the main components of the reaction.
DNA polymerase (green), primers (purple), and dNTPs (pink) are the main components of the reaction.
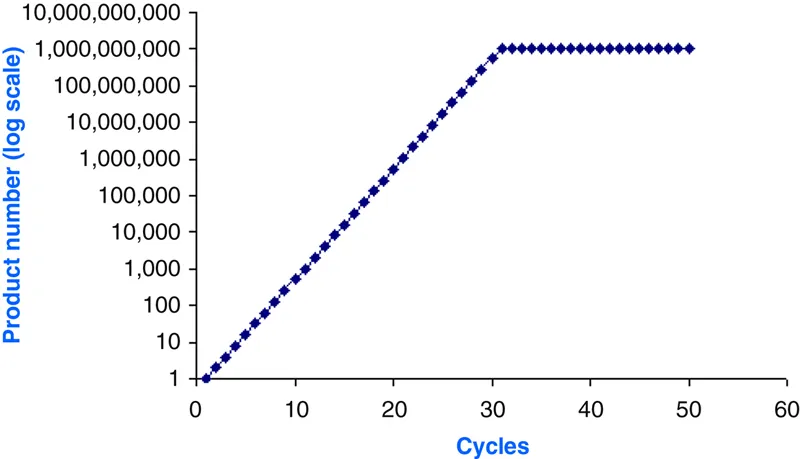
Figure 1.2 A linear plateau phase appears about 30 cycles after the exponential phase.
1.3. PCR components
There are six basic PCR components that are similar among all types of PCRs:
1. Template DNA/RNA: Contains the target sequence to be amplified. (e.g., genomic DNA, plasmid, amplicon, etc.).
2. A pair of primers: Oligonucleotides that define the sequence to be amplified. As the primer concentration increases, the reaction specificity decreases, while efficiency of the reaction increases.

3. Deoxynucleotidetriphosphates (dNTPs): Are DNA-building blocks. As dNTP concentration increases, the reaction specificity and fidelity decreases, while efficiency of the reaction increases.

4. Thermostable DNA polymerase: Is the enzyme that catalyzes the reaction. Originally, the DNA polymerase employed in PCR was isolated from Escherichia coli. However, as PCR involves multiple cycles of temperatures that denature and inactivate the E. coli DNA polymerase, researchers had to manually add a new aliquot of the enzyme at the beginning of each cycle. Fortunately, soon after invention of PCR, a thermostable DNA polymerase, which is nowadays famously known as “Taq,” was isolated from Thermus aquaticus, a bacterium that lives in hot springs. Later, thermostable DNA polymerases were isolated from many other bacterial strains (e.g., Pfu, Tma, and KOD). The optimum temperature for Taq activity is between 75 and 80oC [3] and half-life of this enzyme at 95oC is about 40 min. Taq does not have a proofreading activity (3′–5′ exonuclease), and thus the frequency of mutations incorporated into PCR products is about 1 in 200,000 per nucleotide per duplication of the DNA [4–6]. For applications where fidelity of the DNA polymerase is critical, polymerases such as Pfu and Pwo are used. These DNA polymerases have a proofreading activity (3′–5′ exonuclease) and a fidelity that is about 10 times higher than the fidelity of Taq DNA polymerase [5,6]. For detailed comparison of the thermostable DNA polymerases, see Appendix I.
The concentration of enzymes is defined in units. Each enzyme-supplying company provides the definition of a unit for the enzyme it supplies. For example, New England Biolabs (NEB) defines 1 U of Taq as: the amount of enzyme that incorporates 10 nmols of dNTPs into acid-insoluble material in 30 min at 75oC. Usually, these definitions are close to each other, if one is switching to another company’s enzyme, it is recommended to compare the definitions of units and the concentrations of the supplied en...
Table of contents
- Cover
- Title page
- Table of Contents
- Copyright
- Dedications
- Biography
- Preface
- Acknowledgments
- Note to the Reader
- Chapter 1: Introduction
- Chapter 2: Procedure
- Chapter 3: Good PCR Practices
- Chapter 4: Optimization and Troubleshooting
- Chapter 5: Tips and Tricks
- Chapter 6: Special Cases
- Glossary and Abbreviations
- Appendix A: Agarose Gel Electrophoresis Protocol
- Appendix B: Silver Staining of DNA Gels
- Appendix C: Plasmid Miniprep Protocol (by using “QIAprep Spin Miniprep” Kit)
- Appendix D: Restriction Enzyme Digestion Protocol
- Appendix E: Protocol for DNA Concentration Estimation Using Hoechst 33258
- Appendix F: Formulas
- Appendix G: Common Primers for Sequencing, Promoters, Housekeeping Genes, and Reporters
- Appendix H: Tags
- Appendix I: Thermostable DNA polymerases
- Appendix J: Example of a Daily PCR Log
- Appendix K: Common Buffers and Solutions
- Appendix L: Useful Numbers
- Appendix M: Yeast Colony PCR
- Useful Websites
- References
- Index