![]()
Chapter 1
Introduction
Abstract
Copy number variations (CNVs) currently are most often understood as submicroscopic gains or losses of chromosomal material, either connected with a disease or just one of the many possible genetic variants in man. However decades ago, besides such submicroscopic CNVs, chromosome analysis revealed the existence of cytogenetic visible copy number variations (CG-CNVs). In this chapter a short outline of cytogenetic history is given, highlighting the first detection and overinterpretation and possible meanings of CG-CNVs. Also heterochromatic and euchromatic CG-CNVs are distinguished from submicroscopic CNVs and some specific features of each group are introduced.
Keywords
submicroscopic copy number variations; cytogenetic visible copy number variations (CG-CNVs); heterochromatic CG-CNVS; euchromatic CG-CNVS
Human chromosomes were first visualized in a microscope in the late 1870s [
Arnold, 1879] and were named in 1888 by Heinrich Wilhelm Waldeyer by combining the words
chroma (Greek χρ
μα, meaning color) and soma (Greek σ
μα, meaning body) [
Waldeyer, 1888]. Still, it was another 68 years before the correct modal chromosome number in humans was determined to be 46 [
Tijo and Levan, 1956]. Notably, this particular finding was the starting point of the discipline “human cytogenetics,” which deals with the numerical and structural analysis of human chromosomes. Since that time in 1956, cytogenetics has played a crucial role in pre- and postnatal, as well as tumor cytogenetic diagnostics and research.
Cytogenetics went through different developmental steps, each providing more and better possibilities for the characterization of structurally abnormal and/or supernumerary chromosomes of unknown origin. The history of human cytogenetics can be divided into three major time periods:
• Prebanding (1879–1970)
• Pure banding (1970–1986)
• Molecular cytogenetic era (1986–today), including the recent invention of array-based comparative genomic hybridization (aCGH)
The identification of the first inborn [Lejeune et al., 1959] and acquired chromosomal abnormalities [Nowell and Hungerford, 1960] occurred in the prebanding era. The banding era started with the development of the Q-banding method by Dr. Lore Zech [Schlegelberger, 2013] in 1968 [Caspersson et al., 1968]. Further techniques like C-banding (CBG) or silver staining of the nucleolus organizing regions (NOR) followed in 1971 and 1976, respectively, and completed the cytogenetic set of standard methods for the next decade [Sumner et al., 1971; Bloom and Goodpasture, 1976]. Currently, the GTG-banding approach (G-bands by Trypsin using Giemsa) [Claussen et al., 2002] is still the gold-standard for all cytogenetic techniques. As a result, translocations, inversions, deletions, and insertions can now be detected and described accurately [Pathak, 1979]. The pure banding era ended in 1986 with the first molecular cytogenetic experiment on human chromosomes [Pinkel et al., 1986]. The preferred technique of molecular cytogenetics is fluorescence in situ hybridization (FISH) [Liehr and Claussen, 2002].
The major proceedings in molecular cytogenetics in the past were the comparative genomic hybridization (CGH) [Kallioniemi et al., 1992] and its array-based variant aCGH [Ren et al., 2005]. Also, multicolor-FISH (mFISH) approaches have been developed since 1989 and continue to this day [Liehr et al., 2013]; for example, a FISH-based detection of copy number variants (CNVs), the so-called parental-origin-determination-FISH (pod-FISH) approach, was established recently [Weise et al., 2008]. To which extent next-generation sequencing (NGS) approaches can be helpful to study submicroscopic and microscopically visible CNVs remains to be determined. However, aligning or quantification of long repetitive elements present at different loci of the human genome (like satellite III DNA in all acrocentric short arms and homologues regions in 9p12 and 9q13 [Starke et al., 2002]) is no easy task for any kind of sequencing approach [Sipos et al., 2012].
All the aforementioned (molecular) cytogenetic methods (see also Chapter 6) provide information on the human genome at different levels of resolution [Shinawia and Cheung, 2008]. Even so, already an early finding of cytogenetics was that basically no two clinically healthy individuals are alike on a chromosomal level [Ferguson-Smith et al., 1962; Makino et al., 1966]. Thus, cytogenetic visible copy number variations (CG-CNVs) have been detected and characterized since the early days of cytogenetics. Especially prone to formation of CG-CNVs is the constitutive heterochromatin, defined as “regions that are generally late replicating, rich in repetitive DNA sequences, and genetically inert” [Jalal and Ketterling, 2004], and as
that portion of the genome that remains condensed and intensely stained with DNA intercalating dyes throughout the cell cycle. It represents a significant fraction of most eukaryotic genomes and is generally associated with (…) pericentric regions of chromosomes. Contrary to euchromatin, heterochromatic regions consist predominantly of repetitive DNA, including satellite sequences and middle repetitive sequences related to transposable elements and retroviruses. Although not devoid of genes, these regions are typically gene-poor. Establishment of heterochromatin depends on two basic elements: the histone modification code and the interaction of nonhistone chromosomal proteins. [Rizzi et al., 2004]
In addition to heterochromatic CG-CNVs, more recent findings are euchromatic CG-CNVs and submicroscopic ones. Submicroscopic CNVs are detectable only by molecular cytogenetics or aCGH, and thus here abbreviated as MG-CNVs. Also, there is some overlap of MG-CNVs and CG-CNVs [Manvelyan et al., 2011].
1.1 The Problem
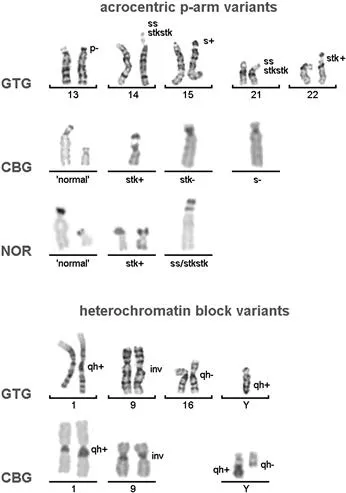
It is well known from banding cytogenetics that some chromosomal regions in the human karyotype are prone to variations more than others; for example, the pericentric regions of chromosomes 1, 9, and 16; the long arm of the Y chromosome; or the short arms of all acrocentrics (Figure 1). More and more regions that might be present in varying copy numbers in the genome without any phenotypic consequences are identified. This accounts not only for CG-CNVs, but even more for the regions of the genome previously considered as junk DNA (i.e., the MG-CNVs) [Schlattl et al., 2011].
Figure 1 Examples for CG-CNVs as seen in routine cytogenetics; nomenclature is according to ISCN 2013. In the two lines where chromosomes are depicted after GTG-banding the left one of two homologous chromosomes represents the normal variant.
Although CG-CNVs are the focus of this book,...