
- 288 pages
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Intracellular Parasitism
About this book
This publication is a collection of essays on the biology of intracellular parasitisms where both bacterial and protozoan parasites are discussed. The juxtaposition of authors representing fields of research emphasizes the many common problems facing intracellular parasites and the hosts that harbor them. In addition, numerous illustrations of how different parasites and host attempt to solve these problems in different ways are provided. The book includes one or more chapters on Bdellovibrio, Chlamydia, Rickettsia, Coxiella, Legionella, Shigellae, Mycobacterium, Microsporidium, Plasmodium, and Toxoplasma. The authors frequently speculate and generalize on the subject matter discussed.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
Chapter 1
POLYPHYLETIC ORIGIN OF BACTERIAL PARASITES
William G. Weisburg
TABLE OF CONTENTS
I. Introduction
A. Bacterial Parasites Have Free-Living Ancestors
B. Gene Sequences as Archeological Records
II. Overview of Bacterial Phylogeny With Particular Attention to Pathogenic Species
A. Eukaryotes, Archaebacteria, and Eubacteria
B. Distribution of Pathogenic and Intracellular Species
III. The Rickettsias
A. Rochalimaea quintana, Agrobacteria, and Plant Pathogenic Species
B. The Tribe Rickettsiae
C. The Family Rickettsiaceae
IV. Do Chlamydiae Have Close Relatives?
V. Evolutionary Steps to Intracellular Parasitism
A. Spectrum of Host-Associated Bacterial Niches
B. Approximately When Did These Various Lineages Begin?
Acknowledgments
References
I. INTRODUCTION
A. Bacterial Parasites Have Free-Living Ancestors
Intracellular parasites offer a unique challenge to biologists, particularly the obligately intracellular bacteria. The challenge lies in that they cannot be studied free of their host; where the organism ends and the host begins can be vague. For this reason, and many others, it is important to consider the intracellular bacteria in an evolutionary context, apart from their present symbiotic state. The knowledge of an obligate parasite’s phylogenetic relatives, as well as a reasonable scenario for evolution into an intracellular niche, are useful for understanding these fastidious bacteria in a manner apart from their hosts.
There are numerous distinctions to consider when describing the intracellular bacteria. Division of the intracellular forms into obligate and facultative species, and division of host-associated bacteria into intracellular and epicellular groups is of dubious use.1 The first dichotomy—obligate vs. facultative—may reflect our ability to culture them. The second pair of terms may be two stages of a dynamic process, for example, an organism has learned how to cope with the outside of a cell, and in (evolutionary) time its descendants may succeed in becoming intracellular. These distinctions are mentioned here because the primary focus of this chapter is to relate, by phylogenetic analysis, the intracellular bacteria to the overall scheme of bacterial evolution, as it is presently understood. The other goal is to speculate on the origin of parasitic bacteria from free-living species. What macroevolutionary steps were taken? When may these events have occurred, and how can these speculations be examined experimentally?
B. Gene Sequences as Archeological Records
The study of bacterial phylogeny has not always been fruitful. The natural classification of species according to their evolutionary diversion and ascent has traditionally relied on grouping together organisms with similar morphological features; for example, Linnaeus used the parts of flowers to classify plants. At the time that bacteria were being taxonomically and systematically ordered, the characteristics available for scrutiny included shape, stain affinity, color, motility, and a few specialized physiological and ecological properties.
In the mid-1960s, Zuckerkandl and Pauling suggested a novel approach to the study of the evolutionary history of species.2 They proposed that the genes of organisms were archeological relics of their ancestors. The sequences of macromolecules behaved as chronometers, and changed at a predictable rate through evolutionary time. If a biologist examined the same gene in a set of species, the two most similar genes would represent the pair of organisms that diverged most recently. In fact, the organisms can be grouped into nested hierarchies and represented as a phylogenetic tree. This revolutionary approach to the study of evolution was particularly far-sighted in view of the crude technology that was available for studying the sequences of genes (nucleic acid sequences) or gene products (proteins). Several technical innovations have occurred since then to allow rapid sequencing of nucleic acids, including RNA fingerprinting, gene cloning, DNA sequencing, RNA sequencing, and DNA oligomer synthesis.
The first workers to apply the principles of Zuckerkandl and Pauling to the study of bacterial evolution were Schwartz and Dayhoff using cytochrome protein sequences,3 and Woese, who examined the sequences of ribosomal RNAs.4,5,6 The ribosomal RNA sequence approach has proven to be more generally applicable. Ribosomes are found in all organisms, and their RNA constituents are approximately homologous in structure and function throughout the biological world.
The ribosomes of bacteria contain three RNA chains, designated 16S (in the small ribosomal subunit) and 23S and 5S (in the larger ribosomal subunit). Sequences of 16S rRNA have been the most widely used for phylogenetic studies. In Escherichia coli it contains 1542 nucleotides.7 In the earliest phylogenetic studies, sequences were partially determined by the oligonucleotide cataloging method.4,5 Beginning in the 1980s, sequence determination of 16S rRNA has been primarily by the primer extension/dideoxynucleotide termination method of Sanger. This sequencing method has been used on two fronts: sequencing cloned rRNA genes, and directly from unfractionated cellular RNA using reverse transcriptase.8,9
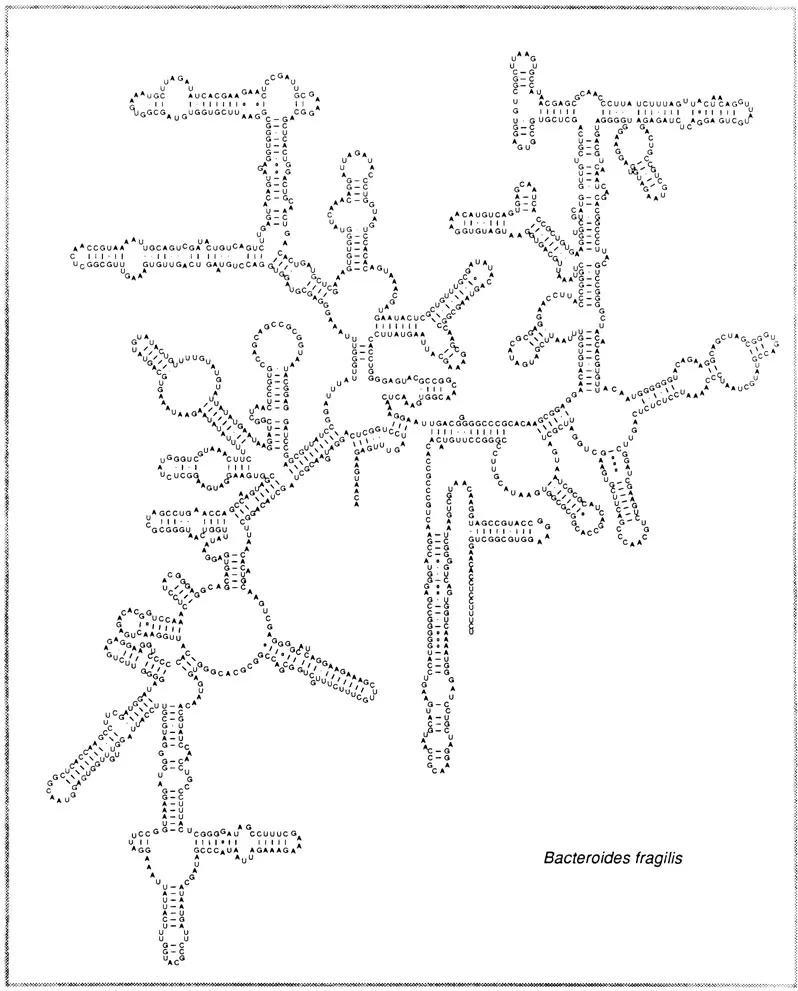
Figure 1 shows the current model for the secondary structure of 16S ribosomal RNA.10,11 Bacteroides fragilis is chosen as a representative eubacterium because of its prominence as an isolate from the human gastrointestinal tract and to exemplify the point that all 16S rRNA secondary structures are approximately identical.11,12 The secondary structure is crucial for phylogenetic study because it serves as the basis for exact alignment of sequences. A group of 16S rRNA sequences are aligned based on the conserved regions as well as on the location of helices and unpaired regions. An alignment then serves as the basis for constructing similarity matrices, distance trees, parsimony analyses, and signature analyses.
The goal of a phylogenetic approach to the comparative study of bacteria is straightforward—to accurately describe the relatedness of species to one another. When the genetic relationships are clear, then one can examine phenotypic, morph...
Table of contents
- Cover
- Title Page
- Copyright Page
- Preface
- The Editors
- Contributors
- Table of Contents
- Chapter 1 Polyphyletic Origin of Bacterial Parasites
- Chapter 2 Cell Envelope Modifications Accompanying Intracellular Growth of Bdellovibrio bacteriovorus
- Chapter 3 The Host Cell/Host Immune Responses and the Intracellular Growth of Chlamydia
- Chapter 4 Antigenic Variation of Chlamydia trachomatis
- Chapter 5 Rickettsia rickettsii: An Enigmatic Pathogen
- Chapter 6 The Rickettsia - Host Interaction
- Chapter 7 A Preview of Rickettsial Gene Structure and Function
- Chapter 8 Persistent Infection with Coxiella burnetii in vitro and in vivo
- Chapter 9 Genetic Diversity of Coxiella burnetii
- Chapter 10 Molecular Strategies for Uptake and Intraphagosomal Growth of Coxiella burnetii in Non-Immune and Immune Hosts
- Chapter 11 The Immunobiology of Legionella pneumophila
- Chapter 12 Intracellular Parasitism of Shigellae
- Chapter 14 Microsporidian Spores as Missile Cells
- Chapter 15 Protein Transport and Membrane Biogenesis in Malaria-Infected Erythrocytes
- Chapter 16 Interactions of Malaria Parasites and Their Host Erythrocytes
- Chapter 17 Sexual and Mosquito Stages of Plasmodium falciparum
- Chapter 18 Active Modification of Host Cell Phagosomes by Toxoplasma gondii
- Chapter 19 Biology of Toxoplasma gondii Host Cell Entry — The Role of Recognition and Attachment for Invasion of Host Cells
- Index
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access Intracellular Parasitism by James W. Moulder in PDF and/or ePUB format, as well as other popular books in Medicine & Medical Microbiology & Parasitology. We have over one million books available in our catalogue for you to explore.