![]()
Design and applications of synthetic information processing circuits in mammalian cells
Jan Lonzarić, Tina Fink and Roman Jerala*
DOI: 10.1039/9781782622789-00001
National Institute of Chemistry, Hajdrihova ulica 19, 1000 Ljubljana, Slovenija. Email:
[email protected]Mammalian cells represent a challenge and opportunity for synthetic biology. The toolbox of regulators implemented in mammalian cells includes small molecule- or physical stimulus-responsive elements from bacteria, yeast and plants, and designed nucleic acid binding proteins such as TALEs and CRISPR/Cas. This enables engineering of mammalian cells to sense versatile external or endogenous signals and process them via designed logic circuits, or rewiring of endogenous signaling pathways for therapeutic responses, demonstrated in disease models of diabetes, inflammation, cancer… Synthetic biology tools are also applied to improve safety and efficiency of engineered therapeutic cells, such as for cancer immunotherapy or stem cells.
1 Introduction
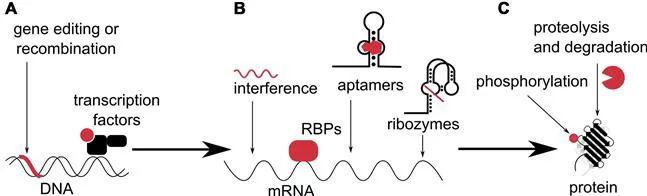
Synthetic biology as a discipline merging life sciences with engineering has since its inception proved to be a valuable tool in research as well as in industrial and medical applications. While wholly synthetic genomes are no longer a fiction,1,2 it is now becoming possible to not only replace an organism's genetic material with synthetic DNA, but to also design the function of that synthetic DNA and produce genetic circuits that either respond to combinations of external or internal signals in therapeutic applications3,4 or provide us with valuable insights into the complex relationships between DNA, RNA and proteins in the functional cell.5,6 The principle idea behind synthetic biology is in fact a simple one—to define functionally separate basic parts that behave in a predictable way and can be joined and recombined as modules into higher system, which will perform much more complex functions, but in turn also act predictably. Basic modules for gene editing, transcriptional control, translational control and functional control of proteins were first developed in prokaryotes, and drawing a parallel to electronic systems, transistors,7 Boolean logic gates,8 oscillators,9 switches,10 band-pass filters11 and many other regulatory modules and devices were assembled. Many of these devices have then been transferred or independently reinvented to function in eukaryotic cells. In this review we will focus on synthetic biology in mammalian cells and aim to present an overview of different mechanism of gene editing and transcription regulation (Fig. 1A) through direct control of transcription factors or through rewired signaling pathways, we will provide a window into construction and function of modules operating on the level of RNA and translation regulation (Fig. 1B), and will describe attempts at construction of faster responsive elements through posttranslational control of proteins and their functions (Fig. 1C). Finally, we will describe examples of interesting and complex devices that represent an important advance towards potentially useful therapeutic applications of mammalian synthetic biology.
Fig. 1Synthetic biology toolbox. Synthetic circuits are regulated on the levels of DNA, RNA and/or proteins. (A) Genes are edited directly or epigenetically regulated with transcription factors. (B) Translation is regulated with RNA interference, RNA-binding proteins (RBPs), aptamers and ribozymes, which can be a part of the mRNA transcript or supplied externally. (C) Proteins are regulated by functional reconstitution, phosphorylation/dephosporylation, proteolysis or degradation/stabilization.
2 Input signals and inducible systems
The first step in designing a controllable synthetic biological circuit is the selection of appropriate input signals. Often, proof of principle circuits are designed so that the input signal is provided by the presence or absence of certain constitutively active elements.12–14 On the other hand, circuits can be designed to conditionally respond to a wide range of externally supplied or intracellular signals, specific to a disease state, physiological conditions or cell type. The latter include the presence of certain DNA elements,15 short RNAs,16 or endogenous proteins,17–19 and circuits can respond to the signals present locally or systemically in an organism, such as metabolites,20–23 neurotransmitters24 or cytokines.25,26 Development of physiologically relevant synthetic sensors is fundamental to development of therapeutic circuits and we will cover some examples of these input systems along with the examples of therapeutic use in a later section of this review. In these opening section however, we will focus on the introduction of synthetic input systems, designed to be orthogonal to the mammalian cell biology.
2.1 Allosterically controlled transcription factors
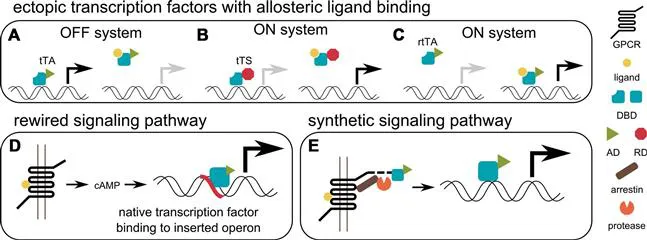
For therapeutic applications of biologic devices as well as for investigation purposes we often want to design systems that respond only to selected input signals without sensing or interfering with the state of the chassis cell or organism in unpredictable ways. This allows us to exert control over the target cells, either to perturb the investigated system or to safely initiate or terminate production of therapeutic effectors. The earliest developed and most well characterized orthogonal sensing systems are based on small chemical inducers. In genetic circuits, the inducers can exert their effect directly on transcription factors (TFs) by allosterically affecting their binding affinity for target DNA.27 Many such transcription factors are derived from bacteria28 and modified to function in mammalian cells by fusion with repression domains (such as the Krüpel associated box—KRAB29) or activation domains (such as the virally derived VP16 domain30 or the human p65 domain31). Additionally, appropriate binding sites for those transcription factors have to be inserted into the promoter region of the gene of interest. The most widely used example of a bacterial transcription factor adapted for mammalian use, and one that superbly illustrates the principles of modular design, is the tet system. The TetR protein, originaly found in bacteria, has first been fused to the VP16 domain to generate a ligand dependent activator that drives expression from a minimal promoter with the tetO binding sites.32 TetR binds to tetO only in the absence of its ligand, doxycyclin, therefore the TetR-VP16 fusion (tTA) creates the so called tet-OFF system that activates transcription in the absence of doxycycline (Fig. 2A). If only transient expression is desired, the tet-OFF system places an inconvenient burden on both the researcher and the cell in having to maintain and tolerate a constantly high concentration of doxycyclin when no protein is being produced. To circumvent this and activate the target gene only in the presence of doxycyclin, two other design options are available—either the fusion of TetR to the KRAB in combination with a constitutively active tetO containing promoter, which could be described as derepression in the absence of the ligand and was termed the TET-dependent transsilencer tTS33 (Fig. 2B), or the reverse tet transcription activator rtTA, which binds to the target DNA only in the presence of the ligand and thereby activates transcription with the help of the VP16 fusion domain34 (Fig. 2C). These options differ in kinetic properties and leakage, which are therefore the key factors that need to be considered during the design of circuits containing the tet systems. Importantly, there is a whole range of bacterial transcription factors responding to different small molecule ligands described, however for most of them mutants that bind to target DNA either in the absence or the presence of ligands are not available, so fusion with either VP16 or KRAB is the preferred strategy in harvesting bacterial transcription factors for the design of OFF and ON systems in mammalian cells, respectively.27 Many other small molecule responsive transcriptional regulators have been harvested from nature, mainly bacteria, such as acetaldehyde responsive elements, which enable cell stimulation through volatile components,35 a urate responsive system, where in addition to the natural sensor addition of a transporter improved the responsiveness,23 and the sensing of the flavonoid phloretin, an apple metabolite, by the regulator from Pseudomonas putida.36
Fig. 2Transduction of information through transcription factors. Many prokaryotic transcription factors are allosterically regulated by small molecule binding. (A) The tet transactivator (tTA) is composed of the TetR DNA-binding domain (DBD) and the VP16 activation domain (AD), which activates transcription in absence of ligand. (B) The tet transsilencer (tTS) is composed of the TetR DBD and a KRAB repression domain (RD), which silences expression in the absence of ligand. (C) The reverse tet transactivator (rtTA) is composed of the reverse tet DBD and the AD VP16, which activates transcription in presence of ligand. (D) A rewired G-protein coupled receptor (GPCR) pathway signals through the native secondary messenger cAMP to activate a native transcription factor, which binds to an ectopically inserted operon. (E) A syntheti...