![]()
| Hsp70 Molecular Chaperones: Versatile Modular Nanomachines that Mediate Multiple Biological Functions |
Lila M. Gierasch*
Abstract
This chapter discusses the multitalented Hsp70 molecular chaperones, with particular emphasis on structural insights into their allosteric mechanism and how this mechanism enables them to perform a panoply of functions in nearly all organisms and cellular compartments. As our focus is the structural aspects of Hsp70 chaperone mechanisms, we call the interested reader’s attention to a number of recent reviews for more extensive discussion of many other topics related to Hsp70s (Clerico and Gierasch, 2014; Mayer, 2010, 2013; Saibil, 2013; Zuiderweg et al., 2013).
The Discovery of the Hsp70 Molecular Chaperone Family
The Hsp70 molecular chaperone family was first described as a set of 70 kDa proteins with greatly increased expression after heat shock, and hence the members were categorized as “heat shock proteins” (Lindquist, 1986; Lindquist and Craig, 1988). It was soon recognized that this family of proteins was widespread in nature, with the identification of Hsp70s from Drosophila (Tissieres et al., 1974) to bacteria (Bardwell and Craig, 1984) to humans (Hunt and Morimoto, 1985). Constitutively expressed homologues were soon discovered in addition to the originally observed heat-inducible Hsp70s, and Hsp70 homologues were found in nearly all eukaryotic cellular compartments, including the cytoplasm, mitochondrion, chloroplast, endoplasmic reticulum (ER), and nucleus (Gething and Sambrook, 1992; McKay, 1993). The association of the Hsp70s with heat shock led to proposed functions in the protection of cells from heat-induced damage, particularly protein unfolding and aggregation. It soon became clear that the Hsp70 family performs a much broader array of cellular functions than simply protecting cells from heat shock damage. For example, the bacterial 70 kDa protein, DnaK, is indeed essential for viability and growth after heat shock (Paek and Walker, 1987), but it is also required for λ phage replication (Georgopoulos, 1977), a role that is based on the facilitation of complex disassembly (Alfano and McMacken, 1989). The ER Hsp70, also known as heavy-chain-binding protein or BiP, was first described as a facilitator of the folding and assembly of immunoglobulins (Munro and Pelham, 1986), but it is also a critical player in protein translocation across the ER membrane (Vogel et al., 1990). In another eukaryotic system, a 70 kDa ATPase is implicated in the disassembly of clathrin coats from endocytic vesicles; originally called the clathrin-uncoating ATPase, it is identical to the constitutively expressed cytoplasmic Hsp70 molecular chaperone, Hsc 70 (Ungewickell, 1985). We now know that Hsp70 family members participate in cellular functions as diverse as early protein biogenesis, disaggregation, blocking misfolding, protection from heat stress, organellar import, disassembly of specific complexes (like clathrin cages), synaptic vesicle exocytosis, translocation into the ER lumen, and more (Clerico and Gierasch, 2014).
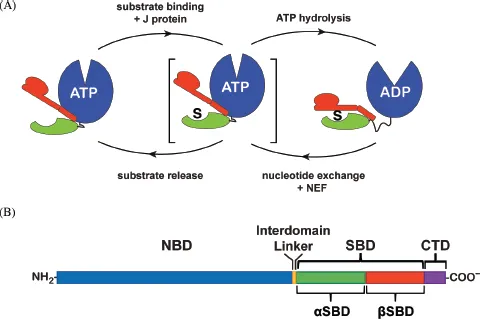
The striking reality is that Hsp70s all share a common biochemical mechanism and somehow this mechanism enables them to perform this array of seemingly disparate functions: essentially, they bind to unstructured regions of their polypeptide substrates with affinity that is modulated by nucleotide binding (McKay, 1993). The physiological context of Hsp70s confers their specialization: both their localization and the team of cochaperones (J-proteins; nucleotide exchange factors, or NEFs) and complicit chaperones they partner with (for example, disaggregases such as ClpB or upstream chaperones such as Hsp90s) permit Hsp70s to adapt to diverse cellular roles. Nonetheless, in all their roles, the underlying allosteric mechanism (Fig. 1.1A) is central: substrates bind with slow on/off-kinetics and high affinity to ADP-bound Hsp70s, and with rapid on/off kinetics and low affinity to ATP-bound Hsp70s (Schmid et al., 1994). Substrate binding to the ATP-bound state, usually facilitated by a J-protein, leads to enhancement of the otherwise low basal Hsp70 ATPase activity, resulting in conversion to the high substrate affinity ADP-bound state. This state is cycled back to the ATP-bound state by the action of a NEF, and the substrate is released and a new one bound. While most of the current mechanistic understanding of Hsp70s derives from studies of E. coli DnaK, it is clear from many studies carried out in a number of laboratories that the same fundamental mechanism underlies the action of all Hsp70s (Clerico et al., 2015; Mayer, 2013; Zuiderweg et al., 2013). The fascinating structural basis for the allosteric mechanism of Hsp70s has only been elucidated quite recently, due to active research in a number of laboratories and a resulting array of structures determined by X-ray crystallography and NMR (Table 1.1). We now appreciate that the ability of Hsp70 chaperones to respond to ligands by a profound structural rearrangement is a result of the intrinsic properties of their constituent domains and is built into these molecular machines by their modular architecture, viz. how these domains are linked to one another. Moreover, this architecture and the resulting energy landscape enable Hsp70 actions to be modulated by key interactions with partner cochaperones.
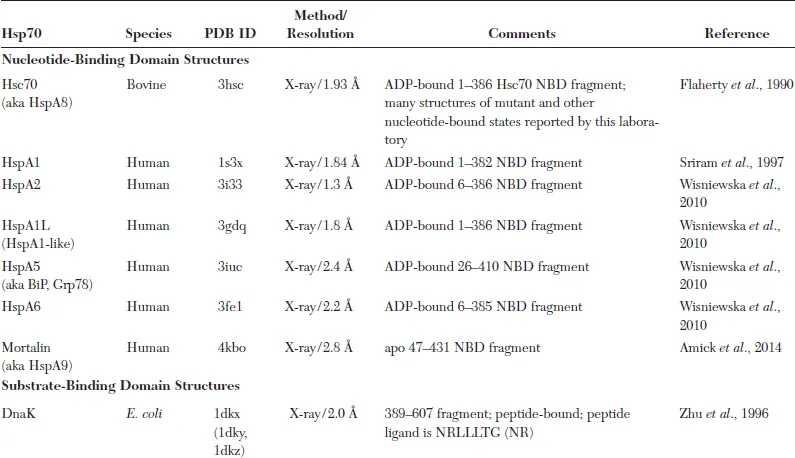
Fig. 1.1 A. Schematic of the allosteric mechanism of Hsp70 molecular chaperones, showing steps that are facilitated by J domain and nucleotide exchange factor (NEF) cochaperones. B. Sequence organization of Hsp70 molecular chaperones. [This figure was prepared by Eugenia Clerico.]
Modular Structural Organization of Hsp70 Molecular Chaperones
The overall structural organization of Hsp70 molecular chaperones is highly conserved, consistent with the common mechanism of action that is at the heart of the multitude of Hsp70 physiological roles: Each family member consists of an N-terminal, 40 kDa nucleotide-binding domain (NBD) connected to a C-terminal approximately 30 kDa substrate-binding domain (SBD) (Fig. 1.1B). As implied by their names, the ligand-binding abilities of Hsp70 chaperones segregate to these domains. Based on its structure, the SBD can be further divided into the βSBD, the first 115 or so residues (390–502; unless otherwise noted, numbering will correspond to the E. coli DnaK sequence), which adopt predominantly β-structure, and αSBD, a helical region of about 100 amino acids (503–607) (see detailed discussion below). The functional collaboration between the NBD and the SBD in Hsp70 chaperones is made possible by an interdomain linker (approximately 12 residues long in prokaryotes and 16 residues long in eukaryotes). The crucial role of the linker in mediating interdomain communication is signaled by several sequence features highly conserved throughout the Hsp70 family, most notably a stretch of four hydrophobic residues, 389–392. A largely unstructured and overall poorly conserved C-terminal domain (CTD) is present in all Hsp70s; this segment is approximately 30 residues long but can vary in length.
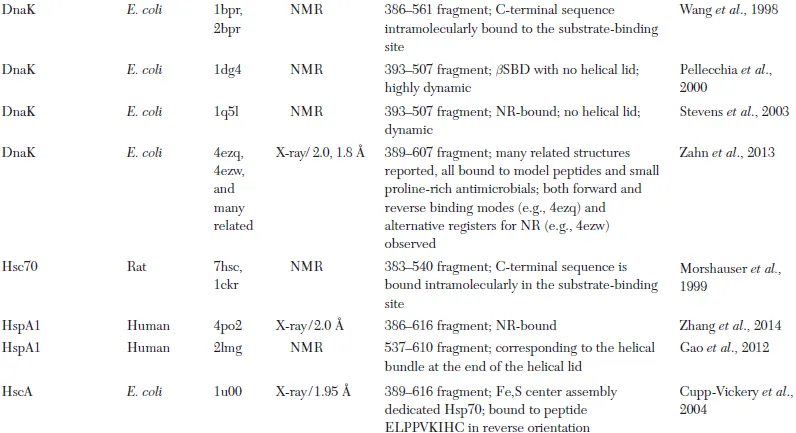
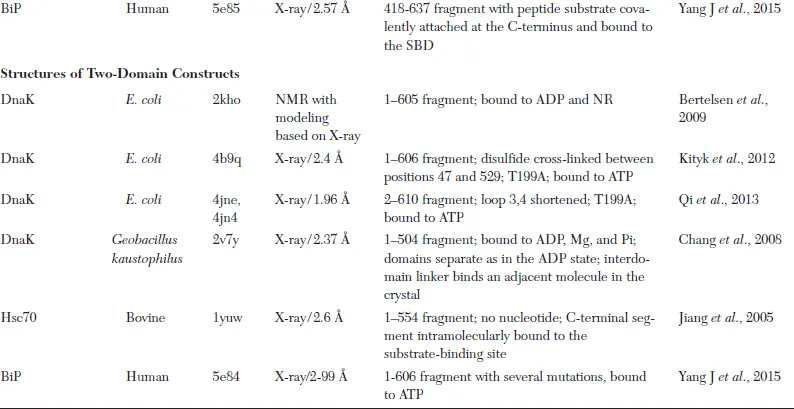
Table 1.1. Representative Atomic Resolution Structures of Hsp70 Chaperones*
* The first structure of a particular Hsp70 domain is listed in the table. Frequently, additional structures have been reported; these are not included unless they added substantially new structural information.
How Does the Modular Structure of Hsp70 Molecular Chaperones Enable Ligand-Mediated Allostery?
The Nucleotide-Binding Domain (NBD)
The X-ray crystal structure of the NBD of Hsc70, the constitutive cytoplasmic Hsp70, was solved soon after the discovery of this chaperone family and has served as a paradigm for the structural features of the Hsp70 NBD. Inspection of the structure immediately pointed to a coarse resemblance to hexokinase, and a stronger homology to actin was soon recognized (Bork et al., 1992; McKay, 1993). These similarities suggested that the Hsp70 NBD had an intrinsic capacity for allosteric conformational change triggered by ligand binding. Like the actin fold, the Hsc70 NBD has a bilobed structure (lobes termed I and II) with two α/β subdomains per lobe (designated IA, IB, IIA, and IIB); the nucleotide-binding site is located deep between the lobes (Fig. 1.2A) (Flaherty et al., 1991). Beneath the nucleotide-binding site are the so-called crossing helices, a canonical feature of the actin fold. The scissors-like crossing helices offer the possibility of gating a hydrophobic binding site by dynamically switching the interhelical angle. In actin, this site is exploited by actin-binding proteins and regulated by allosteric ligands including nucleotide (Dominguez, 2004).
Interestingly, many structures of the Hsc70 NBD were solved for a number of mutant versions and various nucleotide-bound states without seeing any significant conformational change, even between ATP- and ADP-bound states (McKay, 1993). The first hint of actin-like subdomain rearrangements was provided by the structure of DnaK bound to a portion of the NEF GrpE, which showed one subdomain (IIB) twisted with respect to its position in Hsc70 (Harrison et al., 1997). But it required a series of structural breakthroughs in recent years from both NMR and crystallography for the ATP-induced allosteric conformational changes within the NBD, as well as the mode of coupling to the SBD, to be revealed. One breakthrough was provided by an elegant NMR study of the Hsp70 NBD from T. thermophilus comparing ADP-and AMPPNP-bound states, which showed using a residual dipolar coupling analysis that subdomain rotations occurred between these two states (Bhattacharya et al., 2009). The solution of crystal structures of an Hsp70 homologue, Hsp110, in the ATP-bound state (Liu and Hendrickson, 2007), and later two different ATP-bound DnaK forms (Kityk et al., 2012; Liu and Hendrickson, 2007; Qi et al., 2013), confirmed the model that had emerged from the NMR analyses (Bhattacharya et al., 2009), and provided an opportunity for detailed inspection of ADP- and ATP-bound NBDs. Note that because no structure of the ADP-bound DnaK NBD has been reported, the ADP-bound structure of the DnaK NBD is generally modeled based either on that of Hsc70 (Flaherty et al., 1994) or on the structure of the GrpE complex (Harrison et al., 1997). Comparing structures of the NBD bound to ADP (PDB code 3hsc) and ATP (PDB code 4b9q) shows changes in the twist of the two lobes with respect to each other, and both altered angles of the subdomains and altered rotations with respect to each other (Fig. 1.2B). The concerted intradomain conformational change and subdomain rearrangement gates the hydrophobic binding site under the crossing helices and alters the presentation of residues on potential surfaces of interaction. Strikingly, a calculation of normal modes of the NBD fold using the web-based server ElNémo (Suhre and Sanejouand, 2004) revealed that the movements of subdomains between the ADP- and ATP-bound states observed in crystal structures are an intrinsic low frequency motion of the NBD (Zhuravleva and Gierasch, 2011). Similarly, an NMR analysis of 12 different ligand-bound states of the NBD yielded patterns of chemical shift perturbations (Fig. 1.2C) that sho...