![]()
Chapter 1
Nanotechnology and the Unique Role of DNA
Elisa-C. Schöneweiß, Andreas Jaekel and Barbara Saccà∗
Centre for Medical Biotechnology (
ZMB)
University of Duisburg-Essen 45117 Essen, Germany ∗[email protected] This chapter is a clear introduction to the role of DNA molecule in Nanotechnology. A part from describing the canonical and alternative structures, this chapter is aimed at giving fundamentals of self-assembling strategy of DNA nanostructures.
1.1Introduction
“What I want to talk about is the problem of manipulating and controlling things on a small scale.” With these words Richard Feynman introduced his talk at Caltech on December 29th 1959, entitled “There is plenty of Room at the Bottom” [1], which signed the beginning of a new field of science: nanotechnology. In his visionary speech, the Nobel Laureate in Physics anticipated the tremendous possibilities offered by the “staggeringly small world that is below,” some of which became reality only recently. For the first time, scientists were challenged to overstep the minimal size of things that can be manipulated, bringing miniaturization to the smallest possible level. He proposed for example to use light to arrange molecules or even single atoms in a desired pattern on a suitable surface. This idea found later application in a series of top-down approaches, such as photolithography, inject-printing and other micropatterning methods [2–4], all sharing the same principle: the exploitation of the laws of physics to pack a huge amount of information on an exceedingly small space. On the other hand, the opposite process, that is, the extraction of all possible information from a tiny object, was also envisioned as a realistic goal, achievable for example with the development of more powerful microscopes. Nowadays, complex molecular assemblies can be observed almost at atomic resolution thanks to the improvement of electron microscopy [5–7] and kinetic processes can be resolved with picosecond accuracy using advanced single-molecule techniques [8–11].
Storing and using the information carried in a tiny space is the way nature performs its tasks. Fundamental biological processes such as cellular duplication, growth, movement and interactions are all tightly regulated in a microscopic and crowded area. What is mostly striking about it is that the information driving the whole machinery is contained in a few micrometer-large fraction of the cell, the nucleus, in the form of an almost two meter-long chain, the DNA molecule, which carries about one bit of information every 50 atoms. In this sense, the DNA molecule is probably the most brilliant example of nanotechnology in nature, being itself a nanosized object, capable to store and transmit the information necessary to life in a simple and efficient way. It is therefore not surprising that the beginning of the nanotechnology era started only few years after the discovery of the structure and function of DNA by Watson and Crick in 1953 [12, 13] and that latest advancements in the field have been made using DNA as the material.
The idea of employing DNA to construct molecular objects was pioneered by Ned Seeman in the early 1980s [14] and since then structural DNA nanotechnology has rapidly evolved, both in terms of design principles and applications [15]. Using a simple four bases code and applying predictable base pairing rules, this bottom-up approach allows the realization of DNA objects of almost any desired shape and pattern, thus enabling to reach the nanosized world from the molecular level, with sub-nanometer resolution. A notable breakthrough in the field occurred in 2006, when Paul Rothemund published the first scaffolded DNA-origami approach [16]. This method revolutionized our vision of DNA as a construction material, giving accessibility to nanosized objects with a level of sophistication previously only hardly imaginable [17, 18]. Recently other design strategies have emerged [19–22], enlarging the spectrum of attainable structures and demonstrating that the future of DNA design is still bright. This, together with the availability of user-friendly design tools and synthetic accessibility of oligonucleotides at relatively low costs, enabled the exponential growth of DNA nanotechnology in the past few years [23].
Besides solving design challenges, scientists rapidly succeeded in demonstrating the use of those structures for realistic applications, from the development of addressable molecular pegboards for protein patterning [24–27] or encapsulation [28–32], to optoelectronic hybrid materials [33] and organic catalysts [34]. Another field in great expansion is coupled to the advancement of single-molecule technologies, enabling for example the precise localization and counting of molecules in spatially distributed samples or the disclosure of anomalous kinetic events occurring on a time scale normally not accessible by standard methods [8, 35–40]. Recently, DNA nanostructures have also shown interesting properties in cells [41] and are currently explored as promising targeting and delivery systems [42, 43]. For all these applications, the nucleic acid structure must be previously functionalized, i.e. chemically modified with small molecules or proteins [44]. At this purpose, not only classical DNA-conjugation strategies have been implemented and revisited [45], but other methods based on sequence-specificity recognition [46] and protein adaptors [47, 48] have been also successfully applied.
In addition to the use of DNA as a static framework for controlling the spatial arrangement of molecules, the programmability of nucleic acids structures can be conveniently employed to develop dynamic systems, whose interconversion between distinct species occurs in a predictable manner [49–51]. This principle has been successfully employed to develop bioinspired DNA-nanomotors [52, 53] and bioinformatic tools for emulating complex reaction circuits [54].
Finally, one should not forget the enormous contribution to the field of DNA nanotechnology given by Chad Mirkin, who firstly employed the DNA molecule as a recognition motif attached over the surface of metal nanoparticles for biosensing applications [55, 56]. Later, the same principle has been applied for linking distinct nanoparticles together, leading to the generation of colloidal nanocrystals [57, 58] and nanoplasmonic materials with advanced optical properties [59].
In conclusion, although many of the initial Feynman’s predictions became true, one cannot but remaining astonished by the enormous progresses made in the field and the rapid advancement of technologies and design strategies to approach the nanosized world with such a high level of control. In this sense, the DNA molecule represents an ideal candidate for the realization of those self-organizational systems initially envisaged in Feynman’s lecture, which can help us to manipulate matter and understand the basic principles of self-assembly in natural and man-made architectures.
1.2The DNA Molecule
1.2.1The double helical B-form
The fundamental building block of every DNA nanostructure is the DNA double helix. Understanding the structure and chemical–physical properties of the double helix is therefore necessary when the purpose is to design ordered assemblies of single DNA units and using them for further functionalization.
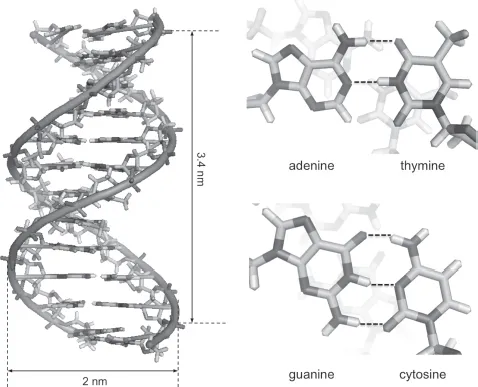
DNA is a polymer composed by the periodic repetition of four possible nucleotides (Figure 1.1). Each nucleotide is made up of three components: a phosphate group, a five-carbon sugar deoxyribose and a nitrogenous base (also termed nucleobase or simply base). The four nucleobases are adenine (A), guanine (G), thymine (T) and cytosine (C). Whereas the sugar–phosphate moieties provide the backbone of the nucleic acid structure, the four bases are responsible for its helical shape. This latter comes mostly from base stacking interactions among the aromatic nitrogenous bases. This is true for both single-stranded and double-stranded DNA. The double-stranded (ds) form, however, requires hydrogen bonding between the bases of each strand in order to form a stable double helix. The process is called base pairing because each nucleobase pairs only with a certain other nucleobase, named “complement”: A pairs with T and G pairs with C. Such a Watson–Crick base-pairing rule is the fundamental law of each DNA design: once given a certain DNA sequence, its complementary sequence is uniquely defined. Such a reliable programmability of DNA nanostructures basically represents the main reason for the rapid and successful implementation of structural DNA nanotechnology.
Figure 1.1. The DNA molecule in its most common B-form is a right-handed helical structure, which is 2 nm wide and rises up ca. 3.4 nm every helical turn (corresponding to ca. 10.5 base pairs (bp)). The four nucleobases (A, G, T, C) and their paring rule are also indicated. Note that whereas AT pairs are stabilized by two h...