![]()
Working on tRNA Structure Project with Alex
Sung-Hou Kim*
Alex was the first person who opened my sense of excitement of scientific discovery which has been driving my scientific curiosity and career ever since. But my encounter with him happened by an “unexpected and chance circumstance.“ In 1966, I was in my last year in the graduate program on crystallographic studies of carbohydrate molecules at the University of Pittsburgh under the guidance of George A. (“Jeff”) Jeffrey, and I decided to learn about “New Biology” (later called Molecular Biology) for my postdoctoral training. One day, Jeff asked me whether I would be interested in doing my postdoctoral research with one of his friends at MIT. When he told me that his friend worked in the Department of Biology, I said “YES” immediately, because I saw an opportunity for learning New Biology, which was my “intention.” At that point I did not know anything about the accomplishments or international reputation of Alex since my graduate study subject was very different from that of Alex’s field. At my first encounter with Alex at MIT, I immediately liked him, especially his friendly openness, relaxed and down-to-earth manner and unpretentiousness, all the attributes that were totally foreign to a young Asian who did his undergraduate and master’s studies in South Korea.
Alex was also the first person from whom I learned that one has to take a risk if one wants to ask a big question. Starting a new project of crystallizing tRNA was such a risk for me, especially since I had no experience in the then new field of macromolecular crystallography. Yet, Alex agreed for me to start the project. However, he “neglected” to mention to me that many laboratories had failed to crystallize tRNA in the past. He probably did not want to scare me off the project or thought that the project needed a fresh, new or even “naïve” approach unencumbered by the knowledge of past failures by many well-known groups. In fact, my first success in tRNA crystallization came out of a naïve approach of using a binary-solvent crystallization system, often used in small molecule crystallization, but never used for macromolecular crystallization because commonly used solvents usually denature macromolecules.
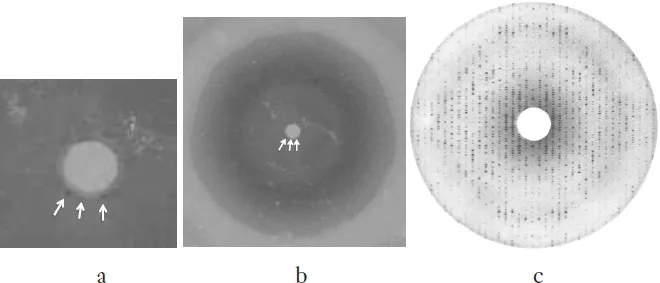
Fig. 1. Three years of search. (a) The very first diffraction pattern of E. coli f-met tRNA taken using a sealed-tube Cu X-ray source. There were no diffraction spots immediately recognizable. But on close examination, there were three barely visible spots at the bottom edge of the shadow of the beam-stop (the white circle in the middle); (b) A magnified view of the beam-stop shadow revealing the three spots indicated by the arrows; (c) 3Å diffraction pattern of yeast phe-tRNA crystals.
The first batch of crystals of E. coli f-met tRNA1 diffracted so poorly that I could barely detect three (what I thought were) diffraction spots at the edge of the shadow of the X-ray beam stop (Fig. 1a, b). It took almost three years of intensive search with “Bud” Suddath and Gary Quigley to finally discover the conditions to reproducibly obtain the crystals that diffracted to about 3Å resolution (Fig. 1c). The three key factors were: changing our sample to then-commercially available yeast phe-tRNA (Boehringer Mannheim), adding spermine hydrochloride, and using vapor-phase equilibrium crystallization system.2
Our tRNA project went into a new phase when I decided to take a faculty position at the Department of Biochemistry, Duke University, in 1972. Alex and I agreed to continue our tRNA project together at MIT and Duke in order to decrease the chances of making mistakes by cross-checking the results of independent interpretations by the two groups. In fact, the first success of such a collaboration came sometime late in 1972, when Bud Suddath and I realized, during a phone conversation comparing the results of chain tracing of a 4Å resolution electron density maps calculated by the two groups, that we were both describing the same backbone structure, which we then published in 1973.3 It revealed an “L” shape, totally unexpected and different from all the predicted models (more than 8) of tRNA structure based on non-crystallographic methods, such as various spectroscopic methods and chemical or UV induced base-modification methods. This discovery was one of the most exhilarating moments for me during the six years of working on this project.
For the all-atom structure of tRNA, the two groups took different approaches to improve the structural models to 3Å resolution: the MIT group used Real Space Refinement method, then commonly used by protein crystallographers, while the Duke group used a new approach, Reciprocal Space Refinement method using group scattering factors, being developed specifically for nucleic acid crystallography at Duke.4 Although most regions of the two models agreed, some details in the loop regions were different. At this point, the two groups became “collaboratively competitive,” but the Duke model prevailed at the end. During this period, the progress report on the MIT model was first published in March, 1974,5 and the all-atom structure of the Duke model was published six months later in August, 1974,6 as the final result of the collaboration between MIT and Duke. Almost at the same time, another all atom structure from a group at the MRC, UK was published, also in August, 1974,7 from 3Å resolution crystals obtained using the same commercially available yeast phe-tRNA, spermine hydrochloride, and vapor-phase equilibrium method, as we had discovered three years earlier2 and used for our final structure.6 The MRC structure also showed the all atom structure to be almost the same as that of the Duke model, giving an independent evidence for the correctness of the structure as well as almost all tertiary base-pairings in the tRNA, which turned out to be common in most tRNAs, for which sequence information was available then.8
Fig. 2. “Image of tRNA,” 2012. A metal sculpture representing the simplified backbone structure of yeast phe-tRNA morphed with the help of the creative metal bending tools and expertise of Byron Rubin, a crystallographer-metal artist.
Some of the most exhilarating moments in my scientific career occurred during the six years (1968–1972 at MIT and 1972–1974 at Duke) of working with Alex and tRNA groups at MIT and Duke (listed in 5 and 6), and the experience of some of such moments has been guiding me to new scientific directions and new questions ever since. Of course, such moments were interleaved with many frustrations, disappointments and doubts, but none of them were overwhelming enough to quench my desire for exhilaration of new discovery. Most of all, the most important benefit I gained from the major events associated with the project was the realization that one or few aspects of human nature played important roles in each event: curiosity, ambition, fame, persistence, competition, generosity, greed, and even politics. Who could have expected that the beauty of the tRNA structure (Fig. 2) harbors so much of my life experiences?
References
1.Kim S-H and Rich A. (1968). Single crystals of transfer RNA: An X-ray diffraction study. Science 162: 1381–1384.
2.Kim S-H, Quigley G, Suddath FL and Rich A. (1971). High-resolution X-ray diffraction patterns of crystalline transfer RNA that show helical regions. Proc. Nat. Acad. Sci. USA 68(4): 841–845.
3.Kim S-H, Quigley GJ, Suddath FL, et al. (1973). Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science 179: 285–288.
4.Sussman JL and Kim S-H. (1976). A preliminary refinement of yeast tRNAPhe at 3Å resolution. In Environmental Effects on Molecular Structure and Properties (Bergman E and B Pullman eds.), 8: 535–545. D. Reidel Publishing Company: Dordrecht-Holland.
5.Suddath FL, Quigley GJ, McPherson A, et al. (1974). Three-dimensional structure of yeast phenylalanine transfer RNA at 3Å resolution. Nature 248: 20–24.
6.Kim S-H, Suddath FL, Quigley GJ, et al. (1974). Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435– 440.
7.Robertus JD, Ladner JE, Finch JT, et al. (1974). Structure of yeast phenylalanine tRNA at 3Å resolution. Nature 250: 546–551.
8.Kim S-H, Sussman JL, Suddath FL, et al. (1974). The general structure of transfer RNA molecules. Proc. Nat. Acad. Sci. USA 71(12): 4970– 4974.
![]()
Crystalline Transfer RNA: The Three-Dimensional Patterson Function at 12-Angstrom Resolution
Sung-Hou Kim and Alexander Rich
Department of Biology, Massachusetts Institute of Technology, Cambridge 02139
Abstract: An orthorhombic form of crystalline formylmethionine transfer RNA has been obtained which contains one molecule as the asymmetric unit of the unit cell. Three-dimensional x-ray diffraction data have been collected up to a resolution of 12 angstroms, and from this a Patterson function has been calculated. The function contains an elongated ridge of interatomic vectors parallel to the c-axis of the crystal. Analysis of the function suggests that the molecules are elongated and dimerized in an overlapping antiparallel fashion along the c-axis. The dimer has a length near 109 angstroms and a width of 35 angstroms in one direction. The individual molecular length is approximately 80 angstroms with an irregular cross section measuring 25 by 35 angstroms.
Transfer RNA (tRNA) is of great importance in the molecular biology of living systems. This molecule can be activated by specific enzymes so that an amino acid is attached to one end. In its activated form tRNA enters the ribosome where it interacts with messenger RNA and participates in the formation of a polypeptide chain during protein synthesis. The molecule thus has a highly specific role in the translation of information from the nucleic acids into the proteins in biological, systems. Because of this central role in the expression of genetic information, there is considerable interest in understanding the three-dimensional configuration of the molecule and the manner in which it carries out its biological functions. Transfer RNA’s have 75 to 85 nucleotides, and the primary sequence of more than a dozen purified species has now been determined (1). This has led to a generally accepted cloverleaf model in which the polynucleotide strand folds back on itself to form short, looped, double-helical portions held together by hydrogen bonding between the purine and pyrimidine bases. If this secondary structure is correct, it is important to unders...