
- English
- ePUB (mobile friendly)
- Available on iOS & Android
eBook - ePub
Human Microbiota and Microbiome, The
About this book
Thousands of different microbial species colonize the human body, and are essential for our survival. This book presents a review of the current understanding of human microbiomes, the functions that they bring to the host, how we can model them, their role in health and disease and the methods used to explore them. Current research into areas such as the long-term effect of antibiotics makes this a subject of considerable interest. This title is essential reading for researchers and students of microbiology.
Tools to learn more effectively

Saving Books

Keyword Search

Annotating Text

Listen to it instead
Information
1 The Stomach and Small and Large Intestinal Microbiomes
Christian U. Riedel,1 Andreas Schwiertz2 and Markus Egert3*
1University of Ulm, Ulm, Germany; 2Institute of Microecology, Herborn, Germany;
3Hochschule Furtwangen University, Campus Villingen-Schwenningen, Germany
1University of Ulm, Ulm, Germany; 2Institute of Microecology, Herborn, Germany;
3Hochschule Furtwangen University, Campus Villingen-Schwenningen, Germany
1.1 Introduction
This introductory chapter provides an updated overview on the composition of the microbiome in the human gastrointestinal tract (GIT); that is, the microbiota of the GIT together with its entire genetic information and the microbe–microbe and host–microbe interactions taking place in this habitat. More specifically, recent scientific advances on the microbiome of the upper (stomach and duodenum) and lower GIT (jejunum, ileum, caecum, colon, rectum), particularly of healthy adults, will be discussed. However, where necessary, some studies performed with diseased patients or animal models will also be presented and integrated into the state-of-the-art-knowledge about the human GIT microbiome. In addition, an update on factors shaping the composition of the GIT microbiome will be given. For a more functional or physiological discussion of the human intestinal microbiome, the reader is referred to Chapter 6, this volume. The structure and function of the microbiome of the uppermost part of the human digestive system, i.e. the oral cavity, are presented and discussed in Chapter 2 of this volume.
From a microbiological point of view, the human GIT can be regarded as the best investigated ecological niche of the human body, although some difficulties exist in obtaining representative samples from various parts of the GIT. Moreover, the human GIT probably represents one of the best investigated microbial ecosystems on earth. This fact can be explained due to the great importance of the GIT microbiota in maintaining and driving human health, disease and well-being: on a quantitative basis, humans can be regarded as a superorganism, consisting of 90% microbial cells and even 99% microbial genes, and the vast majority of the microbial diversity is located in the human GIT (Wilson, 2008). Consequently, the general importance of the GIT microbiome for human health and disease regarding digestion and general metabolism, gut development or immune status is undoubted. Hence, a wealth of literature on the human GIT micobiome is already available, including several current and comprehensive review articles and reviewing book chapters (Wilson, 2008; Doré and Corthier, 2010; Marchesi, 2010; Gerritsen et al., 2011; Walter and Ley, 2011; Willing and Jansson, 2011). For a complementary overview including some of the more classical literature about the human GIT microbiome, the reader is referred to these articles.
1.2 The Microbiota of the Human Stomach
1.2.1 Environmental conditions
The human stomach (Fig. 1.1) is a J-shaped structure with a volume of approximately 1.5 1. It can be differentiated into an upper part (fundus), the main body (corpus) and a lower part (antrum), which is connected to the duodenum part of the small intestine via the pyloric sphincter. The folded stomach epithelium is covered by a protective mucus layer of up to 600 μm thickness. The main functions of the human stomach are temporary food storage, mixture of food and gastric juice to chyme, pre-digestion of proteins by acidic pH and pepsin, and disinfection of the ingested food. The environmental conditions in the stomach are eutrophic – due to ingested food, mucus, desquamated epithelial cells and dead microbes – aerobic and acidic, with a more or less constant temperature of 37°C, i.e. the body temperature of the host. Pronounced daily fluctuations in temperature, pH (from pH 1 to pH 5) and available nutrients are common and linked to ingestions of food and beverages. Bacterial viable counts are strongly dependent on the actual gastric pH and range from 103 to 106/ml (Wilson, 2008; Walter and Ley, 2011).
1.2.2 Composition of the stomach microbiota
Data on the human stomach microbiome are usually collected by investigating biopsies, taken endoscopically after several of hours of fasting. Despite the harsh and antimicrobial environment, recent molecular diversity studies – in particular the widely cited study by Bik and co-workers – have shown, surprisingly, that the human stomach contains a diverse, unevenly distributed microbial community dominated by Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria (Bik et al., 2006). In endoscopic biopsies taken from 23 North American patients with symptomatic upper gastrointestinal disease, they identified 128 phylotypes from 8 phyla by a 16S rRNA gene clone library approach. Several more recent studies corroborated that a remarkable diversity of bacterial genes could be amplified and identified from the human stomach (Andersson et al., 2008; Dicksved et al., 2009; Li et al., 2009; Maldonado-Contreras et al., 2011). While investigating ten Helicobacter pylori-free patients with a Chinese background, Li and co-workers quite clearly corroborated several key findings of the American-based study of Bik and colleagues (Li et al., 2009). With respect to the total number of detected phylotypes (133 versus 127), the number of phyla (8 versus 7) and the most abundant two genera (Streptococcus and Prevotella), both studies yielded strikingly similar results. Anderson and co-workers even detected 262 phylotypes representing 13 phyla in biopsies of the stomach of three H. pylori negative patients with peptic ulcers (Andersson et al., 2008). As a consequence, the human stomach can no longer be considered a mono-associated environment.
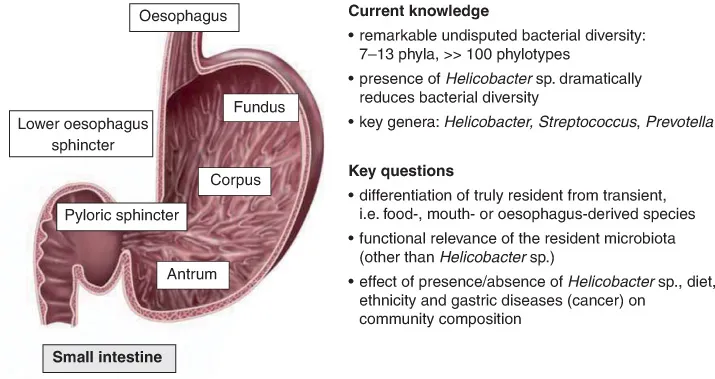
Fig. 1.1. Current knowledge and key questions regarding the microbial ecology of the human stomach.
In the thick mucous layer overlying the gastric epithelium, non-acidophilic bacteria can also be found, in particular H. pylori. When present, H. pylori usually dominates the stomach bacterial community (Andersson et al., 2008). So far, H. pylori is the only bacterium of the human stomach that can be considered unambiguously as a true resident and is considered to contribute to the development of gastritis, peptic ulcers and even gastric cancer (Dorer et al., 2009).
Several recent studies have tried to unravel correlations between the composition of the microbial community in the stomach and the H. pylori status of patients. In a study by Maldonado-Contreras and colleagues, which was focused on patients from developing countries, a positive H. pylori status was correlated with increased relative abundances of (non-Helicobacter) Proteobacteria, Spirochetes and Acidobacteria, while Actinobacteria, Bacteroidetes and Firmicutes were less abundant (Maldonado-Contreras et al., 2011). However, the study also showed that ethnicity had a stronger impact on the stomach community composition than the H. plyori status. Focusing on H. pylori negative patients, Li et al. detected significantly higher abundances of Firmicutes, in particular Streptococcus spp., in the stomach mucosa of patients with antral gastritis (Li et al., 2009). Interestingly, Streptococcus spp., together with bacteria of the genera Lactobacillus, Veillonella and Prevotella, were also abundant members of the stomach community in a study on patients with gastric cancer and a low H. pylori abundance (Dicksved et al., 2009). However, no statistically significant differences were found between the stomach community of cancer and non-cancer patients.
1.2.3 Resident or transient microbiota?
Approximately 1010 microorganisms enter the human stomach every day. As a consequence, a clear differentiation of truly resident from just transient (swallowed) microbial species is difficult. Indeed, the majority of the 33 phylotypes identified in the stomach of all three patients investigated by Andersson et al. were affiliated with the genera Streptococcus, Actinomyces, Prevotella and Gemella, which were also abundant in the throat community (Andersson et al., 2008). However, streptococci were shown to survive in the stomach and to adhere tightly to the mucosa, suggesting they might truly represent resident stomach species (Li et al., 2009). Acid tolerance is clearly a prerequisite for (even just transient) microbial survival in the stomach lumen, and this is why particularly acid-tolerant streptococci, lacto-bacilli, staphylococci and Neisseria spp. have frequently been found in the stomach lumen. It was suggested that some of these bacteria be investigated in more detail for potentially beneficial (probiotic) properties (Ryan et al., 2008). A similar suggestion was recently also put forward for propionibacteria: Delgado and co-workers cultured propionibacteria – mostly affiliated with P. acnes, but devoid of any clear pathogenic properties – from gastric mucosa samples of 8 out of 12 healthy patients and proposed them as true residents of the human stomach (Delgado et al., 2011).
So far, the functional relevance of the surprisingly high microbial diversity in the human stomach is still largely obscure (Lawson and Coyle, 2010). Its elucidation will require more long-term, dynamics-orientated and comparative analyses of mouth, throat and stomach communities and linking of particular physiological conditions, for example those associated with certain gastric diseases and/or the presence/absence of H. pylori, with the composition of the microbiota of the stomach. Eventually, such studies might prepare a basis for the definition of novel therapeutic targets (Lawson and Coyle, 2010).
1.3 The Microbiota of the Small Intestine
1.3.1 Environmental conditions
In the small intestine, the vast majority of food components are digested by mostly host-derived hydrolytic enzymes and subsequently absorbed by the intestinal mucosa. The small intestine can be divided into three major parts (Fig. 1.2), with a more or less constant diameter (~3 cm) but considerable differences in length: i.e. duodenum (~25 cm), jejunum (~1.0 m) and ileum (~2.0 m). The entire epithelium of the small intestine is covered with a thick (up to 250 μm) protective mucus layer, secreted by goblet cells. In order to facilitate digestion and absorption, the surface area of the small intestine is greatly increased to almost 300 m2 by the formation of villi and microvilli (‘brush border’). On transfer through the pyloric sphincter, chyme from the stomach is mixed with intestinal juice (combined excretion of epithelial cells), pancreatic juice and bile by peristaltic movements. Compared to the large intestine, microbial growth is hampered in the small intestine by relatively short food retention times, antimicrobial peptides secreted by paneth cells and bile salts. However, growth conditions for microorganisms improve towards the end of the small intestine. Consequently, the numbers of luminal microorganisms increase from approximately 102 ml–1 in the jejunum up to 108 ml–1 in the terminal ileum (Wilson, 2008; Walter and Ley, 2011).
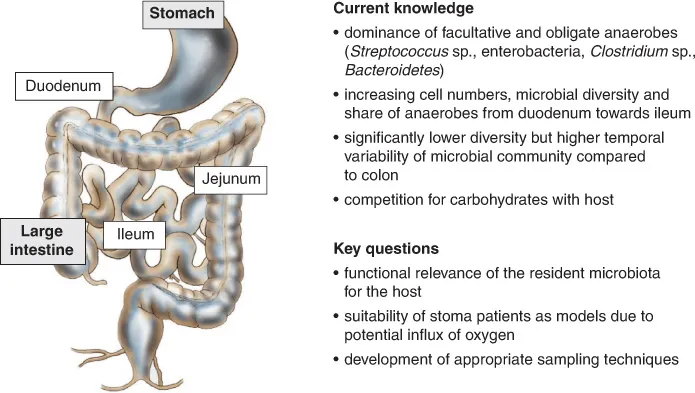
Fig. 1.2. Current knowledge and key questions regarding the microbial ecology of the human small intestine.
1.3.2 Composition of the small intestinal microbiota
Due to its restricted accessibility, the microbiota of the human stomach, and particularly of the small intestine, has been investigated much less intensively than that of the mouth and large intestine or faeces. In particular, data on the small intestinal microbiota of healthy individuals are scarce. Until a few years ago, it was common knowledge that the lumen and mucosa of duodenum and jejunum were colonized at low density by only a few microorganisms, including acid-tolerant streptococci and lactobacilli. Towards the end of the ileum, the lumen was described as being dominated by streptococci, enterococci and coliforms, while in the mucosa, obligate anaerobes (Bacteroides spp., Clostridium spp., Bifidobacterium spp.) could also be found (Wilson, 2008, and studies cited therein). This knowledge has been broadened during the past few years.
In order to characterize the small intestinal microbiota in more detail by molecular means, Booijink and co-workers investigated the ileal effluent of patients with so-called Brooke ileostomies, i.e. patients with an ileum ending in an opening of the abdominal wall, mostly because the colon had to be removed due to colon cancer (Booijink et al., 2010). They showed that the small intestine was characterized by a less diverse and temporarily more fluctuating microbial community than the large intestine (Booijink et al., 2010). Based on community profiles obtained with a phylogenetic microarray, the average community similarity of four patients over 9 days was just 44%. Notably, no Archaea were detected in the effluent samples. Although the community of each patient was highly individual, a hypothetical common ‘core microbiota’ was defined based on these four patients. It comprised bacteria belonging to the genera Clostridium, Enterococcus, Oxalobacter, Streptococcus and Veillonella.
By comparing small intestinal lumen samples obtained from healthy subjects by means of an extended oral catheter with ileal effluent samples, Zoetendal and co-workers very recently showed that the microbial composition of ileal effluent might rather resemble the community in the jejunum (Zoetendal et al., 2012). They identified bacteria belonging to the Bacteroidetes, Clostridium cluster XIVa and Proteobacteria as typical for the ileum. In line with previous studies (Booijink et al., 2010), they corroborated a lower species diversity and significant tempor...
Table of contents
- Cover Page
- Title Page
- Copyright Page
- Contents
- Contributors
- 1 The Stomach and Small and Large Intestinal Microbiomes
- 2 The Oral Microbiome
- 3 The Human Urogenital Microbiome
- 4 The Lung Microbiome
- 5 The Human Skin Microbiome
- 6 Function of the Human Gut Microbiota
- 7 Models of the Human Microbiota and Microbiome In Vitro
- 8 In Vivo and Animal Models of the Human Gut Microbiome
- 9 The Gut Microbiota in Health and Disease
- 10 Next-generation Sequencing Methods to Investigate the Human Microbiome
- 11 Metabonomics for Understanding Gut Microbiome and Host Metabolic Interplay
- Index
- Footnotes
Frequently asked questions
Yes, you can cancel anytime from the Subscription tab in your account settings on the Perlego website. Your subscription will stay active until the end of your current billing period. Learn how to cancel your subscription
No, books cannot be downloaded as external files, such as PDFs, for use outside of Perlego. However, you can download books within the Perlego app for offline reading on mobile or tablet. Learn how to download books offline
Perlego offers two plans: Essential and Complete
- Essential is ideal for learners and professionals who enjoy exploring a wide range of subjects. Access the Essential Library with 800,000+ trusted titles and best-sellers across business, personal growth, and the humanities. Includes unlimited reading time and Standard Read Aloud voice.
- Complete: Perfect for advanced learners and researchers needing full, unrestricted access. Unlock 1.4M+ books across hundreds of subjects, including academic and specialized titles. The Complete Plan also includes advanced features like Premium Read Aloud and Research Assistant.
We are an online textbook subscription service, where you can get access to an entire online library for less than the price of a single book per month. With over 1 million books across 990+ topics, we’ve got you covered! Learn about our mission
Look out for the read-aloud symbol on your next book to see if you can listen to it. The read-aloud tool reads text aloud for you, highlighting the text as it is being read. You can pause it, speed it up and slow it down. Learn more about Read Aloud
Yes! You can use the Perlego app on both iOS and Android devices to read anytime, anywhere — even offline. Perfect for commutes or when you’re on the go.
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Please note we cannot support devices running on iOS 13 and Android 7 or earlier. Learn more about using the app
Yes, you can access Human Microbiota and Microbiome, The by Julian Marchesi, Julian R Marchesi in PDF and/or ePUB format, as well as other popular books in Medicine & Medical Microbiology & Parasitology. We have over one million books available in our catalogue for you to explore.