
Bacterial Resistance to Antibiotics
From Molecules to Man
- English
- ePUB (mobile friendly)
- Available on iOS & Android
About This Book
AN AUTHORITATIVE SURVEY OF CURRENT RESEARCH INTO CLINICALLY USEFUL CONVENTIONAL AND NONCONVENTIONAL ANTIBIOTIC THERAPEUTICS
Pharmaceutically-active antibiotics revolutionized the treatment of infectious diseases, leading to decreased mortality and increased life expectancy. However, recent years have seen an alarming rise in the number and frequency of antibiotic-resistant "Superbugs." The Centers for Disease Control and Prevention (CDC) estimates that over two million antibiotic-resistant infections occur in the United States annually, resulting in approximately 23, 000 deaths.
Despite the danger to public health, a minimal number of new antibiotic drugs are currently in development or in clinical trials by major pharmaceutical companies. To prevent reverting back to the pre-antibiotic era—when diseases caused by parasites or infections were virtually untreatable and frequently resulted in death—new and innovative approaches are needed to combat the increasing resistance of pathogenic bacteria to antibiotics.
Bacterial Resistance to Antibiotics – From Molecules to Man examines the current state and future direction of research into developing clinically-useful next-generation novel antibiotics. An internationally-recognized team of experts cover topics including glycopeptide antibiotic resistance, anti-tuberculosis agents, anti-virulence therapies, tetracyclines, the molecular and structural determinants of resistance, and more.
- Presents a multidisciplinary approach for the optimization of novel antibiotics for maximum potency, minimal toxicity, and appropriated degradability
- Highlights critical aspects that may relieve the problematic medical situation of antibiotic resistance
- Includes an overview of the genetic and molecular mechanisms of antibiotic resistance
- Addresses contemporary issues of global public health and longevity
- Includes full references, author remarks, and color illustrations, graphs, and charts
Bacterial Resistance to Antibiotics – From Molecules to Man is a valuable source of up-to-date information for medical practitioners, researchers, academics, and professionals in public health, pharmaceuticals, microbiology, and related fields.
Frequently asked questions
Information
1
Molecular Mechanisms of Antibiotic Resistance – Part I
1.1 Introduction
- Reducing the concentration of drug able to reach its target, either by preventing its entry or actively removing it from the cell,
- Inactivating or modifying the drug before it reaches the target, either extracellularly or intracellularly,
- Changing the target so that the drug can no longer bind,
- Acquiring an alternative route to carry out the cellular process blocked by the drug.
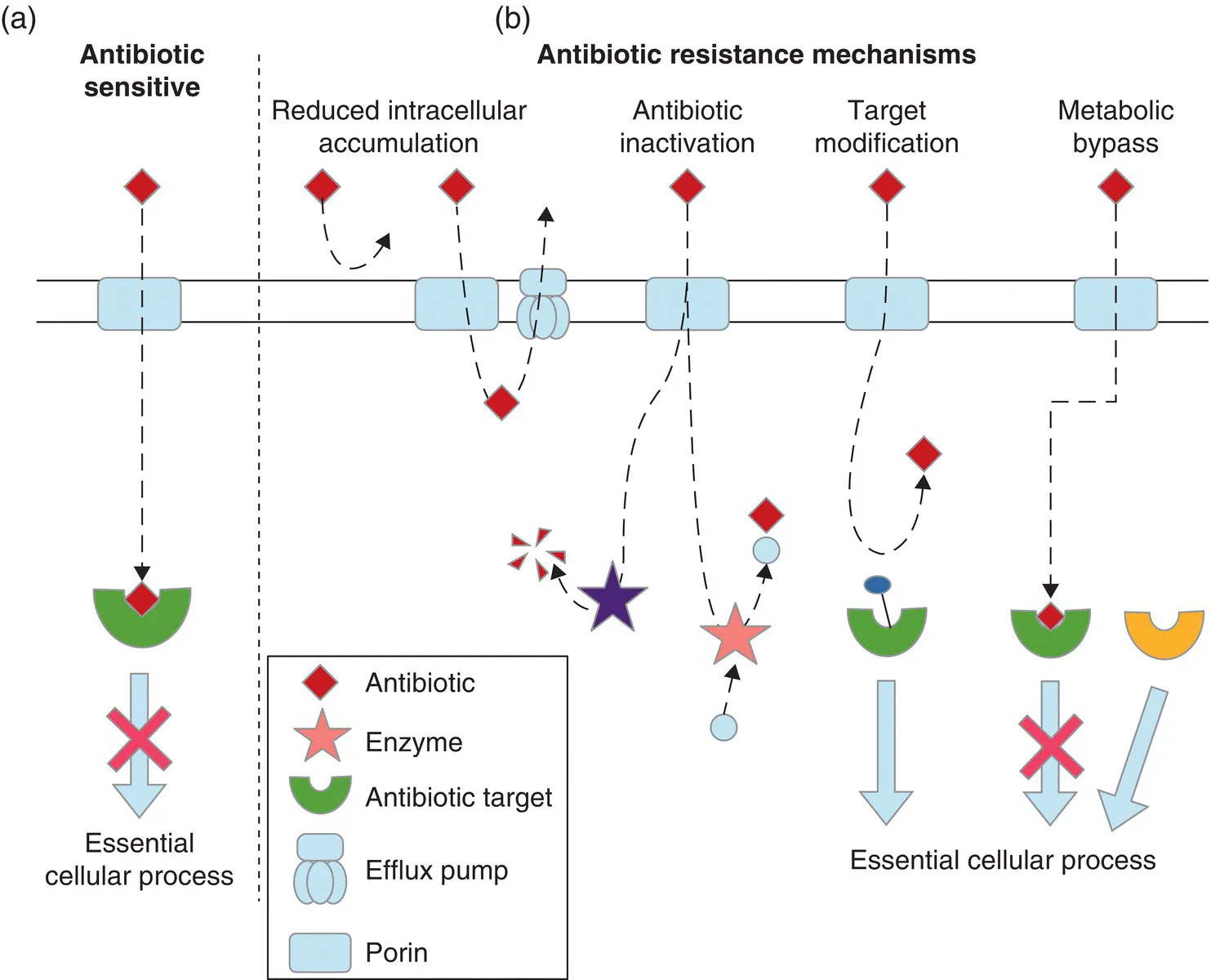
1.2 Molecular Mechanisms of Resistance
1.2.1 Reducing the Intracellular Concentration of a Drug
1.2.1.1 Increased Efflux
- Efflux pumps of the resistance‐nodulation‐division (RND) family are tripartite transporters found in Gram‐negative bacteria, which consist of an inner membrane pump, an outer membrane channel and a periplasmic adaptor protein that connects the two channels. Substrate export is powered by the proton motive force. The best studied example is the AcrAB–TolC pump which was initially discovered in Escherichia coli, but close homologs are widely distributed among Gram‐negative bacteria [14].
- Major facilitator superfamily (MFS) pumps are the largest group of solute transporters and are responsible for most efflux‐mediated resistance in Gram‐positive bacteria [15, 16], although they are also found in Gram‐negative bacteria [17]. They consist of a single polypeptide chain with 12 or 14 membrane spanning domains, with substrate efflux powered by the proton motive force. As an example, several members of this family cause clinically relevant resistance in Staphylococcus aureus. NorA confers resistance to fluoroquinolone antibiotics, QacA exports cationic lipophilic drugs, including biocides such as benzalkonium chloride, and LmrS exports a variety of agents such as lincomycin, linezolid, chloramphenicol and trimethoprim [18–20].
- Small multidrug resistance (SMR) transporters are, as the name suggests, small, having 110–120 amino acid proteins with four membrane spanning domains [15]. They form functional transporters by oligomerizing in the membrane, where they transport substrates using the proton motive force [21]. Examples include QacC from S. aureus and EmrA from E. coli, both of which transport toxic organic cations such as methyl viologen [22, 23].
- Multidrug and toxic compound extrusion (MATE) efflux pumps are commonly found in Gram‐negative bacter...
Table of contents
- Cover
- Table of Contents
- List of Contributors
- Preface
- Foreword
- 1 Molecular Mechanisms of Antibiotic Resistance – Part I
- 2 Molecular Mechanisms of Antibiotic Resistance – Part II
- 3 Resistance to Glycopeptide Antibiotics
- 4 Resistance and Tolerance to Aminoglycosides
- 5 Tetracyclines: Mode of Action and their Bacterial Mechanisms of Resistance
- 6 Fluoroquinolone Resistance
- 7 Dihydropteroate Synthase (Sulfonamides) and Dihydrofolate Reductase Inhibitors
- 8 Anti‐tuberculosis Agents
- 9 Multidrug Resistance
- 10 Anti‐virulence Therapies Through Potentiating ROS in Bacteria
- Index
- End User License Agreement