
- English
- ePUB (mobile friendly)
- Available on iOS & Android
Matrix Metalloproteinase Biology
About This Book
Discussing recent advances in the field of matrix metalloproteinase (MMP) research from a multidisciplinary perspective, Matrix Metalloproteinase Biology is a collection of chapters written by leaders in the field of MMPs. The book focuses on the challenges of understanding the mechanisms substrate degradation by MMPs, as well as how these enzymes are able to degrade large, highly ordered substrates such as collagen. All topics addressed are considered in relation to disease progression including roles in cancer metastasis, rheumatoid arthritis and other inflammatory diseases. The text first provides an overview of MMPs, focusing on the history, the development and failures of small molecule inhibitors in clinical trials, and work with TIMPS, the endogenous inhibitors of MMPs. These introductory chapters establish the foundation for later discussion of the recent progress on the design of different types of inhibitors, including novel antibody based therapeutics. The following section emphasizes research using novel methods to further the study of the MMPs. The third and final section focuses on in vivo research, particularly with respect to cancer models, degradation of the extracellular matrix, and MMP involvement in other disease states.
Written and edited by leaders in the field, Matrix Metalloproteinase Biology addresses the rapidly growth in MMP research, and will be an invaluable resource to advanced students and researchers studying cell and molecular biology.
Frequently asked questions
Information
Chapter 1
Matrix Metalloproteinases: From Structure to Function
1.1 Introduction
1.2 Structures of MMPs
1.2.1 General MMP structure and domain organization
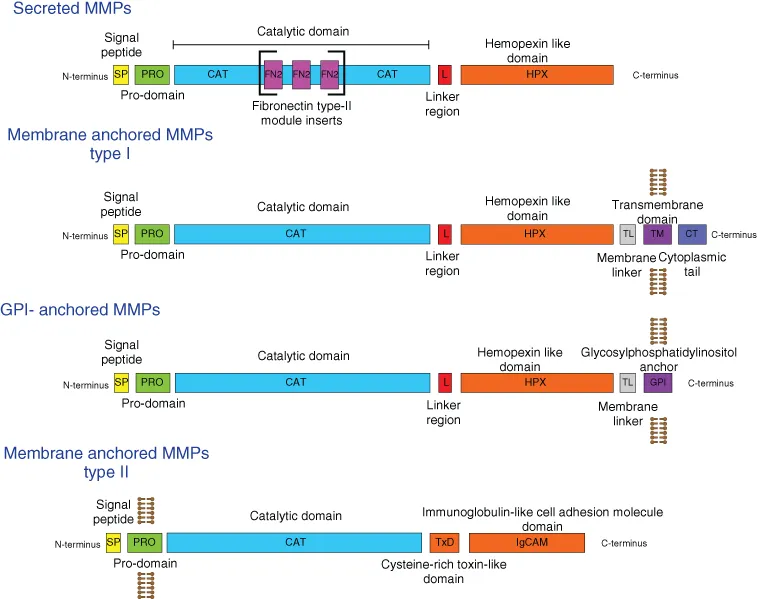
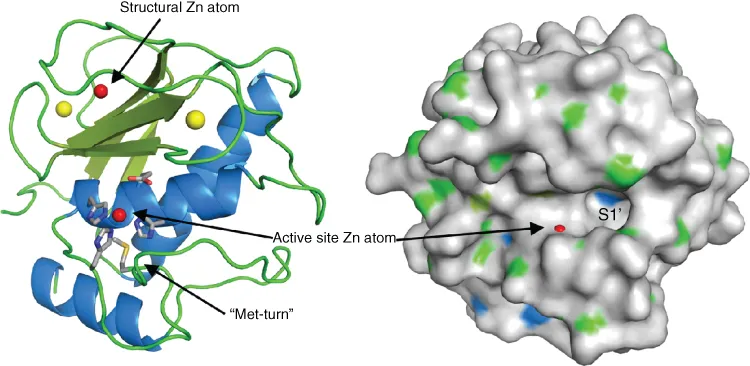
1.2.2 Catalytic domain
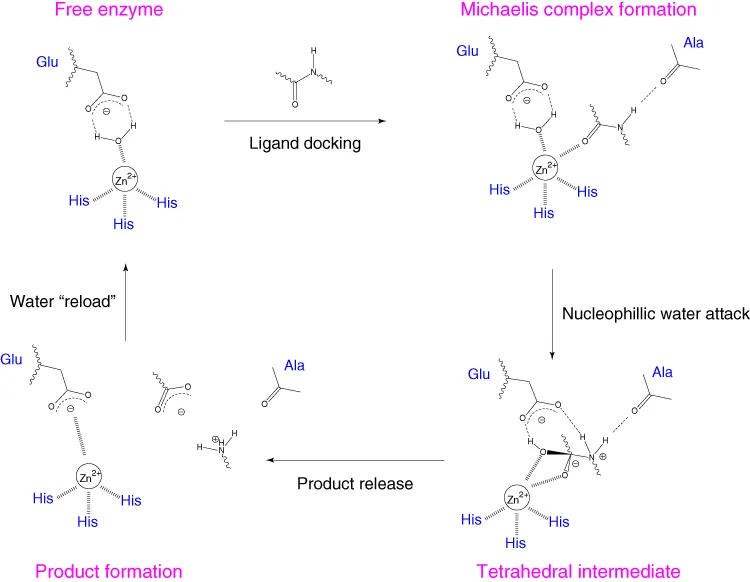
1.2.3 Catalytic mechanism
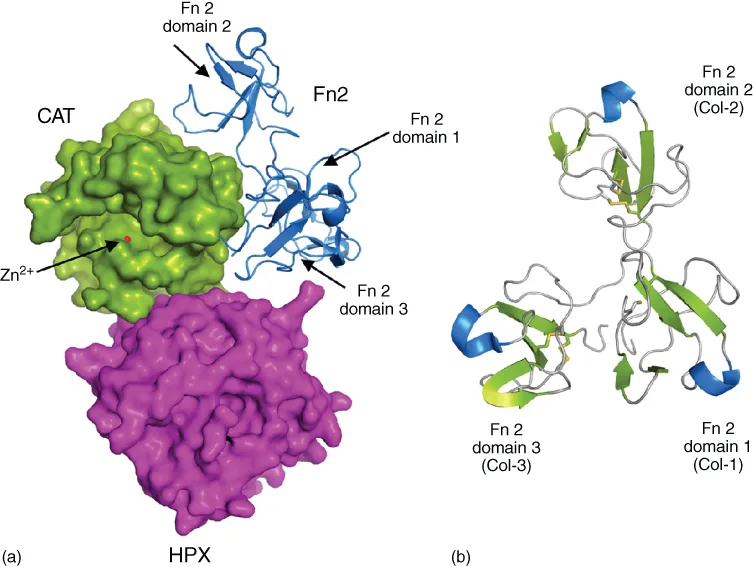
1.2.4 Fibronectin type II-like inserts
1.2.5 Linker region
Table of contents
- Cover
- Title Page
- Copyright
- Table of Contents
- List of Contributors
- Chapter 1: Matrix Metalloproteinases: From Structure to Function
- Chapter 2: Dynamics and Mechanism of Substrate Recognition by Matrix Metalloproteases
- Chapter 3: Matrix Metalloproteinases: From Structure to Function
- Chapter 4: Metzincin Modulators
- Chapter 5: Therapeutics Targeting Matrix Metalloproteinases
- Chapter 6: Matrix Metalloproteinase Modification of Extracellular Matrix-Mediated Signaling
- Chapter 7: Meprin and ADAM Metalloproteases: Two Sides of the Same Coin?
- Chapter 8: Subtracting Matrix out of the Equation: New Key Roles of Matrix Metalloproteinases in Innate Immunity and Disease
- Chapter 9: MMPs: From Genomics to Degradomics
- Chapter 10: MMPs in Biology and Medicine
- Index
- End User License Agreement